Phaser–2.0
Phaser is crystallographic software for phasing macromolecular crystal structures
with maximum likelihood techniques. It is available through the Phenix and CCP4 software
suites, and directly from the authors.
Most people will not need to read this documentation to solve their structure! To solve
a structure by molecular replacement go to Automated
Molecular Replacement and copy and edit
the command script. Similarly, for SAD phasing, go to Automated Experimental Phasing and copy and edit the
command script.
Other good sources of information are found in Frequently Asked Questions and Top Tips
Index to Documentation
1. Introduction
2. General Modes
3. Molecular Replacement Modes
4. Experimental Phasing Modes
5. Keyword Input
6. XML Output (for developers)
7. Python Scripting (for developers)
8. Version History
9. References
1. Introduction
This is the documentation for Phaser–2.0. There
are some changes between this version and previous versions
so input scripts may need editing.
1.1 Modes
Phaser runs in different modes, which perform Phaser's different functionalities.
Modes can either be basic modes or modes that combine the functionality of basic modes.
The mode of operation is controlled with the MODE keyword
| Functionality |
Mode |
Description |
| Anisotropy Correction |
ANO |
Corrects data for anisotropic diffraction, and makes the intensity distribution isotropic |
| Cell Content Analysis |
CCA |
Calculates the expected number of molecular assemblies of given molecular weight in the unit cell using the Matthews coefficient |
| Normal Mode Analysis |
NMA |
Perturbs a structure in rms deviation steps along combinations of normal modes |
| Automated Molecular Replacement |
MR_AUTO |
Combines
anisotropy correction, cell content analysis, fast rotation and
translation functions, and refinement and phasing to automatically
solve a structure by molecular replacement |
| Fast Rotation Function |
MR_FRF |
Anisotropy correction and likelihood-enhanced fast rotation function calculated with Fast Fourier Transform |
| Fast Translation Function |
MR_FTF |
Anisotropy correction and likelihood-enhanced fast translation function calculated with Fast Fourier Transform |
| Brute Rotation Function
| MR_BRF |
Anisotropy correction and full likelihood rotation function calculated in a brute-force search of angles |
| Brute Translation Function |
MR_BTF |
Anisotropy correction and full likelihood translation function calculated in a brute-force search of positions |
| Packing |
MR_PAK |
Tests molecular replacement solutions to see whether they pack into the unit cell without overlap |
| Log-Likelihood Gain |
MR_LLG |
Anisotropy correction and re-scoring of molecular replacement solutions with the full likelihood target function |
| Refinement and Phasing |
MR_RNP |
Anisotropy
correction and optimization of the orientation and position of
molecular replacement models with the full likelihood target function |
| Automated Experimental Phasing |
EP_AUTO |
Combines
anisotropy correction, cell content analysis, and SAD phasing to
automatically solve a structure by experimental phasing |
| SAD Phasing |
EP_SAD |
Refines atoms using the SAD likelihood function, and completes the structure with log-Likelihood gradient maps |
1.2 Keyword Index
1.3 Tutorials and Example Files
The example scripts all refer to the tutorial test cases. The pdb, sequence and
mtz files required to run the tutorials are distributed with Phaser.
- BETA-BLIP
- The crystal structure
of a hetero-dimer of beta-lactamase (BETA) and beta-lactamase inhibitor protein
(BLIP), both with molecular replacement models from crystal structures of
the individual BETA and BLIP components. We thank Mike James and Natalie Strynadka for the diffraction data. Reference:
Strynadka, N.C.J., Jensen, S.E., Alzari, P.M. & James. M.N.G. (1996) Nat. Struct. Biol. 3 290-297.
- Insulin
- The crystal structure of insulin phased on intrinsic sulphurs. We thank Paul Adams for the diffraction data.
Reference: Adams (2001) Acta Cryst D57. 990-995.
1.4 Bug Reports
2. General Modes
2.1 Anisotropy Correction
MODE
ANO corrects the experimental data for anisotropy. Data (amplitude
and associated sigma) are corrected for anisotropy and output to FILEROOT.mtz
with column label set to the input column label with the addition of _ISO.
Example command script to correct BETA-BLIP data for anisotropy
beta_blip_ano.com
phaser ‹‹
eof
MODE ANO
TITLe beta blip data correction
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ROOT beta_blip_ano # not the default
eof
Compulsory Keywords
Optional Keywords
2.2 Cell Content Analysis
MODE
CCA determines the composition of the crystals using the "new"
Matthews coefficients of Kantardjieff & Rupp (2003) "Matthews coefficient
probabilities: Improved estimates for unit cell contents of proteins, DNA
and protein-nucleic acid complex crystals". Protein Science 12:1865-1871.
The molecular weight of ONE complex or assembly
to be packed into the asymmetric unit is given with the COMPosition
keyword, and the possible Z values (number of copies of the complex or assembly)
that will fit in the asymmetric unit and the relative frequency of their
corresponding VM values is reported. RESOlution
should be set to the maximum resolution that has been observed for the crystal.
Example script for cell content analysis for BETA-BLIP
beta_cca.com
phaser ‹‹
eof
TITLe BETA-BLIP cell content analysis
MODE CCA
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
RESO 3.0
ROOT beta_blip_cca # not the default
eof
Compulsory Keywords
Optional Keywords
2.3 Normal Mode Analysis
MODE
NMA writes out pdb files that have been perturbed along normal
modes, in a procedure similar to that described by Suhre & Sanejouand
(Acta Cryst. D60, 796-799, 2004). Each run of the program
writes out a matrix FILEROOT.mat
that contains the eigenvectors and eigenvalues of the atomic Hessian, and
can be read into subsequent runs of the same job, to speed up the analysis.
Do normal mode analysis only, write out eigenfile but not coordinates
beta_nma.com
phaser ‹‹
eof
TITLe beta normal mode analysis
MODE NMA
ENSEmble beta PDB beta.pdb IDENtity 100
XYZOut OFF
ROOT beta_nma # not the default
eof
Write out pdb files perturbed in 0.5 �ngstrom rms intervals in "forward" (positive dq values) along modes
7 and 10 (and combinations of 7 and 10)
beta_nma_pdb.com
phaser ‹‹
eof
TITLe beta normal mode analysis pdb file generation
MODE NMA
ENSEmble beta PDB beta.pdb IDENtity 100
ROOT beta_nma_pdb # not the default
EIGEn beta_nma.mat
NMAPdb MODE 7 MODE 10 RMS 0.5 FORWARD
eof
Compulsory Keywords
Optional Keywords
3. Molecular Replacement Modes
Phaser should be able to solve most structures with the Automated
Molecular Replacement mode, and this is the first mode that you should try.
Give Phaser your data (How to Define Data) and your
models (How to Define Models), tell Phaser what to
search for (use SEARch
keyword), and a list of possible spacegroups (in the same pointgroup - use the
SGALternative
keyword). The flow diagram for the automated molecular replacement mode is shown
below. If this doesn't work (see "Has Phaser Solved It?"),
you can try selecting peaks of lower significance
in the rotation function in case the real orientation was not within the selection
criteria. By default peaks above 75% of the top peak are selected (see "How
to Select Peaks"). See "What to do in difficult
cases" for more hints and tips. If the automated molecular replacement
mode doesn't work even with non-default input you need to run the modes of Phaser
separately. The possibilities are endless - you can even try exhaustive searches
(translations of all orientations) if you want - but experience has shown that
most structures that can be solved by Phaser can be solved by relatively simple
strategies.
3.1 Automated Molecular Replacement
MODE
MR_AUTO combines the anisotropy correction, likelihood enhanced
fast rotation function, likelihood enhanced fast translation function, packing
and refinement modes for multiple search models and a set of possible spacegroups
to automatically solve a structure by molecular replacement. Top solutions
are output to the files FILEROOT.sol,
FILEROOT.#.mtz and FILEROOT.#.pdb
(where "#" refers to the sorted solution number, 1 being the best, and only 1 is output by default). Many structures
can be solved by running an automated molecular replacement search with
defaults, giving the ensembles that you expect to be easiest to find first.
Example command script for finding BETA and BLIP. This is the minimum input,
using all defaults (except the ROOT filename).
beta_blip_auto.com
phaser ‹‹
eof
TITLe beta blip automatic
MODE MR_AUTO
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
SEARch ENSEmble beta NUM 1
SEARch ENSEmble blip NUM 1
ROOT beta_blip_auto # not the default
eof
Example command script for finding BETA and BLIP. The spacegroup recorded
on the mtz file is P3221 but the other hand is also a possibility.
Both search orders (BETA first, BLIP second and BLIP first, BETA second)
are tried, using the PERMutations ON keyword. We would not normally recommend
using the PERMutations ON keyword for this case, as it is obvious that the
larger molecule should be easier to find first. To speed up the calculation
only the top peak after the translation function is taken into refinement.
beta_blip_auto_sg.com
phaser ‹‹
eof
TITLe beta blip automatic
MODE MR_AUTO
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
SEARch ENSEmble beta NUM 1
SEARch ENSEmble blip NUM 1
PERMutations ON # not the default
SGALternative HAND # not the default
ROOT beta_blip_auto_sg # not the default
FINA TRA SELEct NUM 1 # not the default
eof
Compulsory Keywords
Optional Keywords
Flow Diagram for Automated Molecular Replacement in Phaser

3.2 Has Phaser Solved It?
Ideally, only the number of solutions you are expecting should be found.
However if the signal-to-noise of your search is low, there will be noise peaks in the final selection also.
A highly compact summary of the history of a solution is given in the annotation of
a solution in the .sol file.
This is a good place to start your analysis of the output. The
annotation gives the Z-score of the solution at each rotation and
translation function, the number of clashes in the packing, and the
refined LLG. You should see the TFZ (the translation function Z-score)
is high at least for the final components of the solution, and that the
LLG (log-likelihood gain) increases as each component of the solution
is added. For example, in the case of beta-blip the annotation for the
single solution output in the .sol file shows these features
SOLU SET RFZ=11.0 TFZ=22.6 PAK=0 LLG=434 RFZ=6.2 TFZ=28.9 PAK=0 LLG=986 LLG=986
SOLU 6DIM ENSE beta EULER 200.920 41.240 183.776 FRAC -0.49641 -0.15752 -0.28125
SOLU 6DIM ENSE blip EULER 43.873 80.949 117.141 FRAC -0.12290 0.29306 -0.09193
You should always at least glance through the summary of the logfile. One thing
to look for, in particular, is whether any translation solutions with a
high Z-score have been rejected by the packing step, especially
with a small number of clashes. Such a solution may be correct, and the
clashes may arise only because of differences in small surface loops. If
this happens, repeat the run allowing a suitable number of clashes with
the PACK keyword. Note that, unless there is specific evidence in the logfile that
a high TF-function Z-score solution is being rejected with a few clashes, it is much better
to edit the model to remove the loops than to increase the number of allowed clashes. Packing criteria
are a very powerful constraint on the translation function, and increasing the number of allowed clashes
beyond a few (e.g. 1-5) will increase the search time enormously without the possibility of generating any
correct solutions that would not have otherwise been found.
For a rotation function, the correct
solution may be in the list with a Z-score under 4, and will not be found
until a translation function is performed and picks out the correct solution.
For a translation function the correct solution will generally have a Z-score
(number of standard deviations above the mean value) over 5 and be well
separated from the rest of the solutions.
Of course, there will always be exceptions! Note, in particular, that in
the presence of translational NCS, pairs of similarly-oriented molecules
separated by the correct translation vector will give large Z-scores, even
if they are incorrect, because they explain the systematic variation in
intensities caused by the translational NCS.
| TF Z-score |
Have I solved it? |
| less than 5 |
no |
| 5 - 6 |
unlikely |
| 6 - 7 |
possibly |
| 7 - 8 |
probably |
| more than 8 |
definitely |
3.3 What to do in difficult cases
Not every structure can be solved by molecular replacement, but the right
strategy can push the limits. What to do when the default jobs fail depends
on why your structure is difficult.
3.3.1 Flexible Structure
The relative orientations of the domains may be different in your crystal
than in the model. If that may be the case, break the model into separate
PDB files containing rigid-body units, enter these as separate ensembles,
and search for them separately. If you find a convincing solution for one
domain, but fail to find a solution for the next domain, you can take advantage
of the knowledge that its orientation is likely to be similar to that of
the first domain. The ROTAte AROUnd
option of the brute rotation search can be used to restrict the search to
orientations within, say, 30 degrees of that of the known domain. Allow
for close approach of the domains by increasing the allowed clashes with
the PACK
keyword by, say, 1 for each domain break that you introduce.
Alternatively, you could try generating a series of models perturbed by
normal modes, with the NMAPdb keyword. One of these
may duplicate the hinge motion and provide a good single model.
3.3.2 Poor or Incomplete Model
Signal-to-noise is reduced by coordinate errors or incompleteness of the
model. Since the rotation search has lower signal to begin with than the
translation search, it is usually more severely affected. For this reason,
it can be very useful to use the subsequent translation search as a way
to choose among many (say 1000) orientations. Try increasing the number
of clustered orientations in an AUTO job using the keyword FINAL,
e.g. FINAL ROT SELEct PERCent 65.
If that fails, try turning off the clustering feature in the save step (FINAL ROT CLUSter OFF),
because the correct orientation may sit on the shoulder of a peak in the
rotation function.
As shown convincingly by Schwarzenbacher et al. (Schwarzenbacher,
Godzik, Grzechnik & Jaroszewski, Acta Cryst. D60, 1229-1236,
2004), judicious editing can make a significant difference in the quality
of a distant model. In a number of tests with their data on models below
30% sequence identity, we have found that Phaser works best with a "mixed
model" (non-identical sidechains longer than Ser replaced by Ser).
In agreement with their results, the best models are generally derived using
more sophisticated alignment protocols, such as their FFAS protocol.
If there are clear peaks in the self-rotation function, you can expect orientations
to be related by this known NCS. Methods to automatically use such information
will be implemented in a future version of Phaser. In the meantime, you can
work out for yourself the orientations that would be consistent with NCS and
use the ROTAte AROUnd
option to sample similar orientations. Alternatively, you may have an oligomeric
model and expect similar NCS in the crystal. First search with the oligomeric
model; if this fails, search with a monomer. If that succeeds, you can again
use the ROTAte AROUnd
option to force a subsequent monomer to adopt an orientation similar to the
one you expect.
It is frequently the case that crystallographic and non-crystallographic rotational
symmetry axes are parallel. The combination generates translational NCS, in
which more than one unique copy of the molecule is found in the same orientation
in the crystal. This can be recognized by the presence of large non-origin
peaks in the native Patterson map. If one copy of the search model can be
found, then the translational NCS tells you where to place another copy. Unfortunately,
the presence of translational NCS can make it difficult to solve a structure
using Phaser, because the current likelihood targets do not account for the
statistical effects of NCS. If there is a small difference in the orientation
of the two molecules (which will show up as a reduction in the height of the
non-origin Patterson peak as the resolution is increased), it may help to
use data to higher resolution than the default, because the translational
NCS is partially broken.
The
automated mode of Phaser is fast when Phaser finds a high Z-score
solution to your problem. When Phaser cannot find a solution with a
significant Z-score, it "thrashes", meaning it maintains a list of
100-1000's of low Z-score potential solutions and tries to improve
them. This can lead to exceptionally long Phaser runs (over a week of
CPU time). Such runs are possible because the highly automated script
allows many consecutive MR jobs to be run without you having to
manually set 100-1000's of jobs running and keep track of the results.
"Thrashing" generally does not produce a solution: solutions generally
appear relatively quickly or not at all. It is more useful to go back
and analyse your models and your data to see where improvements can be
made. Your system manager will appreciate you terminating these jobs.
It is also
not a good idea to effectively remove the packing test by setting the
allowed number of clashes with the packing test to a high number (e.g
PACK 1000). Unless there is specific evidence in the logfile that a
high TF-function Z-score solution is being rejected with a few clashes,
it is much better to edit the model to remove the loops than to
increase the number of allowed clashes. Packing criteria are a very
powerful constraint on the translation function, and increasing the
number of allowed clashes beyond a few (e.g. 1-5) will increase the
search time enormously without the possibility of generating any
correct solutions that would not have otherwise been found.
Phaser is has powerful input, output and
scripting facilities that allow a large number of possibilities for
altering default behaviour and forcing Phaser to do what you think it
should. However, you will need to read the information in the manual
below to take advantage of these facilities!
3.4 How to Define Data
You need to tell Phaser the name of the mtz file containing your data and
the columns in the mtz file to be used using the HKLIn
and LABIn
keywords. Additional keywords (BINS
CELL OUTLier RESOlution
SPACegroup) define how the data are used.
Phaser must be given the models that it will use for molecular replacement.
A molecular replacement model is constructed in one of two ways - either
by making an ensemble from a set of aligned homologous structures, entered
as pdb files, or by entering a model from a map, entered as structure factors
in an mtz file. Each ensemble is treated as a separate type of rigid body
to be placed in the molecular replacement solution. An ensemble should only
be defined once, even if there are several copies of the molecule in the
asymmetric unit.
Fundamental to the way in which Phaser uses MR models (either from coordinates or maps) is to estimate
how the accuracy of the model falls off as a function of resolution, represented by the Sigma(A) curve.
To generate the Sigma(A) curve, Phaser needs to know the
RMS coordinate error expected for the model and the fraction of the scattering power in the asymmetric unit
that this model contributes.
If fp is the fraction scattering and RMS is the rms coordinate
error, then
Sigma(A) = SQRT{fp*[1-fsol*exp(-Bsol*(sin(theta)/lambda)2)]}
* exp{-(8 Pi2/3)*RMS2*(sin(theta)/lambda)2}
where fsol(default=0.95) and Bsol(default=300�2) account for the effects
of disordered solvent on the completeness of the model at low resolution.
Molecular replacement models are defined with the ENSEmble
keyword and the COMPosition
keyword. The ENSEmble
keyword gives (amongst other things) the RMS deviation for the Sigma(A) curve.
The COMPosition
keyword is used to deduce the fraction of the scattering power in the asymmetric unit
that each ensemble contributes. The composition of the asymmetric unit is defined either by entering the
molecular weights or sequences of the components in the asymmetric unit,
and giving the number of copies of each. Expert users can also enter the
fraction of the scattering of each component directly, although the composition
must still be entered for the absolute scale calculation.
- You have one structure as a model with 44% sequence identity to the
protein in the crystal.
- ENSEmble
mol1 PDB homology1.pdb IDENtity
.44
- You have three structures as models with 44%, 39% and 35% identity to
the protein in the crystal.
- ENSEmble
mol2 PDB
homology1.pdb IDENtity .44 PDB
homology2.pdb IDENtity .39 PDB
homology3.pdb IDENtity .35
- You have an NMR Ensemble as a model. There is no need to split the coordinates
in the pdb file provided that the models are separated by MODEL and ENDMDL
cards. In this case the homology is not a good indication of the similarity
of the structural coordinates to the target structure. You should use
the RMS option; several test cases have succeeded where the ID was close to 100% with an RMS value of
about 1.5� (see table below).
- ENSEmble
mol3 PDB nmr.pdb RMS
1.5
The RMS deviation is determined directly from RMS
or indirectly from IDENtity
in the ENSEmble
keyword using the formula RMS = max(0.8,0.4*exp(1.87*(1.0-ID))) where ID is the fraction identity.
The RMS deviation estimated from ID may be an underestimate of the true value if there is a slight conformational
change between the model and target structures. To find a solution in these cases it may be necessary to increase the
RMS from the default value generated from the ID, by say 0.5 Angstroms. The table below can be used as a
guide as to the default RMS value corresponding to ID.
| Sequence ID |
RMS deviation |
| 100% |
0.80� |
| 64% |
0.80� |
| 63% |
0.799� |
| 50% |
1.02� |
| 40% |
1.23� |
| 30% |
1.48� |
| 20% |
1.78� |
| --> limit 0% |
2.60� |
If you construct a model by homology modelling, remember that the RMS error
you expect is essentially the error you expect from the template structure (if not worse!).
So specify the sequence identity of the template, not of the homology model.
- You have low resolution electron density of your model. This density
has been cut out and converted to structure factors in a large cell.
- ENSEmble
mol1 HKLIn mol1.mtz F
= Fmol1 P = Pmol1 EXTEnt
23 25 29 RMS 2.0 CENTre
4 3 30 PROTein MW 10241 NUCLeic
MW 0
When using density as a model, it is necessary to specify both the extent
(x,y,z limits) of the cut-out region of density, and the centre of this
region. With coordinates, Phaser can work this out by itself. This information
is needed, for instance, to decide how large rotational steps can be in
the rotation search and to carry out the molecular transform interpolation
correctly. In the case of electron density, the RMS value does not have
the same physical meaning that it has when the model is specified by atomic
coordinates, but it is used to judge how the accuracy of the calculated
structure factors drops off with resolution. A suitable value for RMS can
be obtained, in the case of density from an experimentally-phased map, by
choosing a value that makes the SigmaA curve fall
off with resolution similar to the mean figures-of-merit. In the case of
density from an EM image reconstruction, the RMS value should make the SigmaA
curve fall off similar to a Fourier correlation curve used to judge the
resolution of the EM image.
For detailed information, including tutorial with example scripts, see
Using density as a model
The composition is the total amount of protein and nucleic acid that you have in the asymmetric unit not the fraction of the asymmetric unit that you are searching for!
So if you have a model for only one of two components in the asymmetric
unit, you need to enter the molecular weight of both components as the
composition, or if you are searching for only one copy of 12 in the
asymmetric unit, you need to multiply the molecular weight of the
single copy by 12 for the composition.
You can mix compositions entered by molecular weight with those entered by sequence.
- You have one protein (with MW 21022) in the asymmetric unit
- COMPosition PROTein MW 21022
- You have three copies of a protein (with MW 21022) in the asymmetric unit
- COMPosition PROTein MW 21022
- COMPosition PROTein MW 21022
- COMPosition PROTein MW 21022
- Another way of entering the same thing is
- COMPosition PROTein MW 21022
NUMber 3
- Yet another way of entering the same thing is
- COMPosition PROTein MW 63066
- You have two copies of a protein (with MW 21022), two copies of a protein
(with MW 9843) and RNA with (MW 32004) in the asymmetric unit
- COMPosition PROTein MW 21022
NUMber 2
- COMPosition PROTein MW 9843
NUMber 2
- COMPosition NUCLeic MW 32004
The composition is the total amount of protein and nucleic acid that you have in the asymmetric unit not the fraction of the asymmetric unit that you are searching for!
So if you have a model for only one of two components in the asymmetric
unit, you need to enter the sequence of both components as the
composition, or if you are searching for only one copy of 12 in the
asymmetric unit, you need to multiply the number of copies of the
sequence of the single copy by 12 for the composition.
You can mix compositions entered by molecular weight with those entered by sequence.
- You have one protein (with sequence in fasta format in the file prot1.seq) in the asymmetric unit
- COMPosition PROTein SEQuence prot1.seq
- You have three copies of a protein (with sequence in fasta format in the file prot1.seq) in the asymmetric unit
- COMPosition PROTein SEQuence prot1.seq
- COMPosition PROTein SEQuence prot1.seq
- COMPosition PROTein SEQuence prot1.seq
- Another way of entering the same thing is
- COMPosition PROTein SEQuence prot1.seq
NUMber 3
- Yet another way of entering the same thing is to make a sequence file
with all the amino acids concatenated together (prot1.seq3)
- COMPosition PROTein SEQuence prot1.seq3
- You have two copies of a protein (with sequence in fasta format in
the file prot1.seq), two copies of a protein (with sequence in fasta format
in the file prot2.seq) and RNA with (with sequence in fasta format in
the file nucl1.seq) in the asymmetric unit
- COMPosition PROTein SEQuence prot1.seq
NUMber 2
- COMPosition PROTein SEQuence prot2.seq
NUMber 2
- COMPosition NUCLeic SEQuence nucl1.seq
- Each copy of Ensemble mol1 gives 22% of the scattering
- COMPosition
ENSEmble mol1 FRACtional
0.22
- Each copy of Ensemble mol2 gives 78% of the scattering
- COMPosition
ENSEmble mol2 FRACtional
0.78
Phaser writes out files ending in ".sol"
and ".rlist" that
contain the solution information from the job. The root of the files is given
by the ROOT
keyword. By default, the root filename is PHASER. These files can be read
back into subsequent runs of Phaser to build up solutions containing more
than one molecule in the asymmetric unit.
"PHASER.sol" files
are generated by all modes (rotation function modes with VERBOSE output),
and contain the current idea of potential molecular
replacement solutions.
"PHASER.rlist"
files are generated by the rotation function modes, and are for performing
translation functions. (They are also produced by degenerate (2D) translation
functions, for performing a translation function to find the third dimension)
To include the files you should use the preprocessor
command @
@ filename.sol
@ filename.rlist
For simple MR cases you don't really need to know how to define molecular replacement solutions.
However, for difficult cases you might need to edit the files "PHASER.sol"
and "PHASER.rlist"
files manually
3.6.1 "sol" Files
At different stages of molecular replacement, an Ensemble will be oriented
but not positioned (after the rotation search), or oriented and positioned
(after the translation search), or, rarely, oriented and the position in 2
of 3 dimensions known. These three states correspond to solutions defined
by the keywords SOLUtion
3DIM, SOLUtion
6DIM, and SOLUtion
5DIM. Each Ensemble in the asymmetric unit has its own SOLUtion
keyword. Solutions of the type
3DIM are given by the rotation function, solutions of the type
6DIM are given by the translation function, and solutions of the type
5DIM are given by the degenerate translation function. Examples are:
- One copy of mol1 with known orientation and position (fractional coordinates)
- SOLUtion 6DIM
ENSEmble mol1 EULEr
17 20 32 FRACtional 0.12 0.05 0.74
- One copy of mol1 with known orientation only
- SOLUtion 3DIM
ENSEmble mol1 EULEr
17 20 32
- One copy of mol1 with known orientation and only the coordinates in 2 dimensions is
known. The degenerate direction is defined as the direction perpendicular
to the plane in which the position is given.
- SOLUtion 5DIM ENSEmble mol1 EULEr 17 20 32 DEGEnerate X FRACtional 0.05 0.74
When more than one (potential) molecular replacement solution is present, the solutions
are separated with the SOLUTION
SET keywords. For example, if the rotation function and
translation function for mol1 were very clear, then there will only be one
type of 6DIM solution for mol1.
If the rotation and translation functions for mol2 were then not clear, there
will be a series of possible 6DIM
solutions for mol2.
- SOLUtion SET
- SOLUtion 6DIM
ENSEmble mol1 EULEr
17 20 32 FRACtional 0.12 0.05 0.74
- SOLUtion 6DIM
ENSEmble mol2 EULEr
5 183 230 FRACtional 0.71 0.54 0.81
- SOLUtion SET
- SOLUtion 6DIM
ENSEmble mol1 EULEr
17 20 32 FRACtional 0.12 0.05 0.74
- SOLUtion 6DIM
ENSEmble mol2 EULEr
51 93 75 FRACtional 0.08 0.57 0.25
Useful Tip
If you have the coordinates of a partial solution with the pdb
coordinates of the known structure in the correct orientation and
position, then you can force Phaser to use these coordinates by
manually creating a .sol file of the following form and including it in
the Phaser command script with the @filename preprocessor command (or including it directly in the script)
- SOLUtion SET
- SOLUtion 6DIM
ENSEmble mol1 EULEr
0 0 0 FRACtional 0 0 0
3.6.2 "rlist" Files
These files define a rotation function list. The peak list is given with a
series of SOLUtion
TRIAl keywords.
SOLUtion TRIAl ENSEmble
mol1 EULEr 17 20 32 SCORE 4.5
SOLUtion TRIAl ENSEmble
mol1 EULEr 67 65 51 SCORE 4.4
SOLUtion TRIAl ENSEmble
mol1 EULEr 67 112 81 SCORE 4.3
If a partial solution is already known, then the information for the currently
"known" parts of the asymmetric unit is given in the form used for
the PHASER.sol file, followed
by the list of trial orientations for which a translation function is to be
performed.
SOLUtion SET
SOLUtion 6DIM ENSEmble
mol1 EULEr 17 20 32 FRACtional
0.12 0.05 0.74
SOLUtion TRIAl ENSEmble
mol1 EULEr 44 20 32 SCORE 5.8
SOLUtion TRIAl ENSEmble
mol1 EULEr 67 65 51 SCORE 5.2
SOLUtion SET
SOLUtion 6DIM ENSEmble
mol1 EULEr 17 20 32 FRACtional 0.13 0.55 0.76
SOLUtion TRIAl ENSEmble
mol1 EULEr 83 9 180 SCORE 6.3
SOLUtion TRIAl ENSEmble
mol1 EULEr 8 36 92 SCORE 4.2
SOLUtion TRIAl ENSEmble
mol1 EULEr 48 87 10 SCORE 4.0
If a degenerate translation function is performed, then a SOLUtion
TRIAl line is produced with the degenerate translation information
present, ready for performing the translation function on the third dimension.
SOLUtion TRIAl ENSEmble
mol1 EULEr 17 20 32 DEGEnerate
X FRACtional 0.05 0.74
3.7 How to Control Output
The output of Phaser can be controlled with the following optional keywords.
The ROOT keyword
is not compulsory (the default root filename is "PHASER"),
but should always be given, so that your jobs have separate and meaningful
output filenames.
Optional Keywords
Where HKLOut ON
is given as an optional keyword, Phaser produces an mtz file with "SigmaA"
type weighted Fourier map coefficients for producing electron density maps
for rebuilding.
| MTZ Column Labels |
Description |
| FWT |
PHWT |
Amplitude and phase for 2m|Fobs|-D|Fcalc| exp(i alpha-calc) map |
| DELFWT |
PHDELWT |
Amplitude and phase for m|Fobs|-D|Fcalc| exp(i alpha-calc) map |
| FOM |
m, analogous to the "Sim" weight, to estimate the reliability of alpha-calc |
3.8 How to Select Peaks
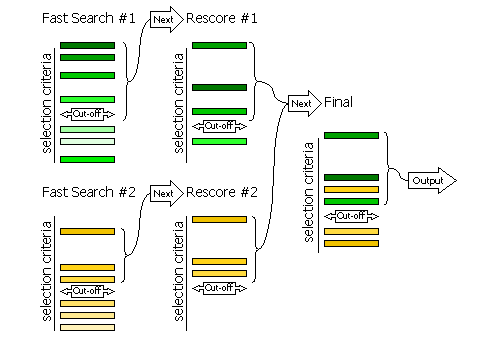
3.8.1 Fast Searches With Rescoring
The selection of peaks for the fast rotation and fast translation function
with rescoring of the top peaks with the full likelihood
target (default RESCORE ON),
is done in three steps, controlled by the keyword
FINAL
For automated molecular replacement, specifying [ROT|TRA] after
the keyword determines which of the rotation or translation function the selection
criteria apply to. If neither is specified, the selection criteria apply to both.
| Keyword |
Applies |
| FINAL [ROT|TRA] STEP 1 |
Controls the selection of peaks from the fast
search that will be rescored with the full likelihood target
|
| FINAL [ROT|TRA] STEP 2 |
Controls the selection of peaks from the rescoring to be
combined with other searches (e.g translation functions of different rotations, or
rotation functions with different fixed components present)
|
| FINAL [ROT|TRA] STEP 3 |
Controls the selection of peaks from the merged list for final output
|
Diagram showing how peaks are selected in three stages
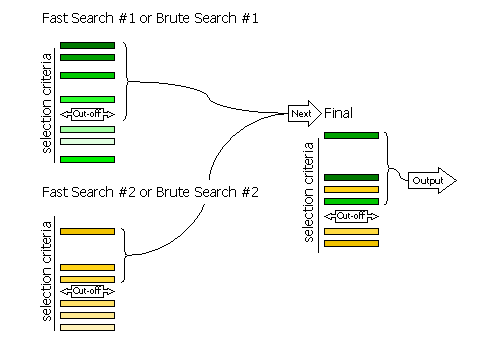
3.8.2 Fast Searches Without Rescoring and Brute Searches
If RESCORE OFF is
requested (no rescoring of the fast search peaks is performed), or if the
brute rotation or translation searches are carried out, then there
are only two stages to selection: selection of peaks from the individual searches,
and selection of peaks from the combined list of solutions.
Selection of peaks at each stage is controlled, respectively, by the keywords
FINAL [ROT|TRA] STEP 2 and
FINAL [ROT|TRA] STEP 3.
Diagram showing how peaks are selected in two stages
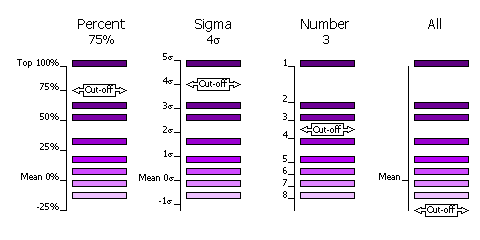
3.8.3 Criteria
The selection of peaks saved for output in the rotation and translation functions
can be done in four different ways.
| Sub-Keyword |
Description |
Use |
| FINAL [ROT|TRA] STEP [1|2|3] SELEct PERCent <CUTOFF> |
Percentage of the top peak, where the value of the top peak is defined as
100% and the value of the mean is defined as 0%. |
Default, cutoff=75%. This criteria has the advantange that at least one peak (the top peak) always survives the selection.
If the top solution is clear, then only the one solution will be output, but if the distribution of
peaks is rather flat, then many peaks will be output for testing in the next
part of the MR procedure (e.g. many peaks selected from
the rotation function for testing with a translation function). |
| FINAL [ROT|TRA] STEP [1|2|3] SELEct SIGma <CUTOFF> |
Number of standard deviations (sigmas) over the mean (the Z-score) |
Absolute significance test. Not all searches will produce output if the
cutoff value is too high (e.g. 5 sigma). |
| FINAL [ROT|TRA] STEP [1|2|3] SELEct NUMber <CUTOFF> |
Number of top peaks to select |
If the distribution is very flat then it might be better to select a fixed
large number (e.g. 1000) of top rotation peaks for testing in the translation function. |
| FINAL [ROT|TRA] STEP [1|2|3] SELEct ALL |
None: all peaks are selected |
Enables full 6 dimensional searches, where all the solutions from the rotation
function are output for testing in the translation function. This
should never be necessary; it would be much faster and probably just as likely
to work if the top 1000 peaks were used in this way. |
Diagram showing selection criteria
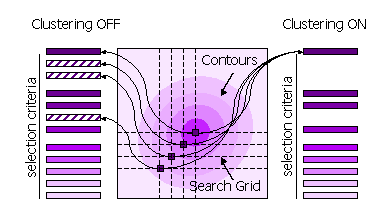
3.8.4 Clustering
Peaks can also be clustered or not clustered prior to selection in steps 1 and 2.
| Sub-Keyword |
Description |
Use |
| FINAL [ROT|TRA] STEP [1|2] CLUSTER OFF |
All high peaks on the search grid are selected |
Default for STEP 1, because the position of the maximum may be different when the fast score
and the full likelihood function are used. |
| FINAL [ROT|TRA] STEP [1|2] CLUSTER ON |
Points on the search grid with higher neighboring points are removed from the selection |
Default for STEP 2. |
Diagram showing clustering
3.9 Basic Modes for Molecular Replacement
3.9.1 Fast Rotation Function
MODE
MR_FRF combines the anisotropy correction and likelihood-enhanced
fast rotation function (2), optionally rescored
with the full rotation likelihood function (1),
to find the orientation of a model in molecular replacement. Top rotation
solutions are output to the file FILEROOT.rlist
for input to a translation function. Top rotation solutions are also output
to the file FILEROOT.sol.
Example command script for fast rotation function to find the orientation
of BETA.
beta_frf.com
phaser ‹‹
eof
TITLe beta FRF
MODE MR_FRF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
SEARCH ENSEmble beta
ROOT beta_frf
eof
Example command script for fast rotation function to find the orientation
of BLIP knowing the position and orientation of BETA, with the position
and orientation of BETA input from the command line.
blip_frf_with_beta.com
phaser ‹‹
eof
TITLe blip FRF with beta rotation and translation
MODE MR_FRF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 #beta
COMPosition PROTein MW 17522 #blip
SEARch ENSEmble blip
SOLUtion 6DIM ENSEmble beta EULEr 201 41 184 FRACtional -0.49408 -0.15571
-0.28148
ROOT blip_frf_with_beta
eof
Example command script for fast rotation function to find the orientation
of BLIP knowing only the orientation of BETA, with the orientation of BETA
input using the output solution file from the beta_frf.com
job above.
blip_frf_with_beta_rot.com
phaser ‹‹
eof
TITLe blip FRF with beta R
MODE MR_FRF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
SEARch ENSEmble blip
@beta_frf.sol # solution file output by phaser
ROOT blip_frf_with_beta_rot
eof
Compulsory Keywords
Optional Keywords
3.9.2 Brute Rotation Function
MODE
MR_BRF combines the anisotropy correction and brute force likelihood
rotation function (1) to find the orientation
of a model in molecular replacement. Top rotation solutions are output to
the file FILEROOT.rlist for
input to a translation function. Top rotation solutions are also output
to the file FILEROOT.sol.
Example command script for brute rotation function to find the orientation
of BETA
beta_brf.com
phaser ‹‹
eof
TITLe beta BRF
MODE MR_BRF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
SEARch ENSEmble beta
ROOT beta_brf
eof
Example command script for brute rotation function to find the optimal orientation
of BETA in a restricted search range and on a fine grid around the position
from the fast rotation search.
beta_brf_around.com
phaser ‹‹
eof
TITLe beta BRF fine sampling
MODE MR_BRF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
SEARch ENSEmble beta
ROTAte AROUnd EULEr 201 41 184 RANGE 10
SAMPling ROTation 0.5
XYZOut ON # not the default
TOPFiles 1 # not the default
ROOT beta_brf_around
eof
Compulsory Keywords
Optional Keywords
3.9.3 Fast Translation Function
MODE
MR_FTF combines the anisotropy correction and likelihood-enhanced
fast translation function (3), optionally rescored
by the full likelihood translation function (1),
to find the position of a previously oriented model in molecular replacement.
Top translation solutions are output to the file FILEROOT.sol.
Example command script for finding the position of BETA after the rotation
function has been run and the results output to the file beta_frf.rlist
beta_ftf.com
phaser ‹‹
eof
TITLe beta FTF
MODE MR_FTF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
@beta_frf.rlist
ROOT beta_ftf
eof
Example command script for finding the position of BLIP after the rotation
function has been run and the results output to the file blip_frf_with_beta.rlist,
which has the SOLUtion 6DIM keyword
input for BETA and the SOLUtion
TRIAL keyword input for the orientations to try for BLIP with the
translation function.
blip_ftf_with_beta.com
phaser ‹‹
eof
TITLe beta FTF
MODE MR_FTF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
@blip_frf_with_beta.rlist
ROOT blip_ftf_with_beta
eof
Compulsory Keywords
Optional Keywords
3.9.4 Brute Translation Function
MODE
MR_BTF combines the anisotropy correction and brute force likelihood
translation function (1) to find the position
of a previously oriented model in molecular replacement. Top translation
solutions are output to the file FILEROOT.sol.
Example command script for brute Translation function to find the position
of BETA after the rotation function has been run
beta_btf.com
phaser ‹‹
eof
TITLe beta BTF
MODE MR_BTF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
@beta_frf.rlist
TRANslate AROUnd FRACtional POINt -0.49408 -0.15571 -0.28148 RANGe 5
ROOT beta_btf
eof
Example command script for brute Translation function to find the position
of BETA degenerate in X after the rotation function has been run
beta_btf_degen_x.com
phaser ‹‹
eof
TITLe beta degenerate X
MODE MR_BTF
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
@beta_frf.rlist
TRANslate DEGEnerate X
ROOT beta_btf_degen_x
eof
Compulsory Keywords
Optional Keywords
3.9.5 Refinement and Phasing
MODE
MR_RNP combines the anisotropy correction and refinement against
the likelihood function (1) to optimize full or
partial molecular replacement solutions and phase the data. At the end of
refinement, the list of solutions is checked for duplicates, which are pruned.
Refined solutions are output to the file FILEROOT.sol.
Example command script to refine a set of solutions
beta_blip_rnp.com
phaser ‹‹
eof
TITLe beta blip rigid body refinement
MODE MR_RNP
HKLIn beta_blip.mtz
LABIn F=Fobs SIGF=Sigma
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
ROOT beta_blip_rnp # not the default
HKLOut OFF # not the default
XYZOut OFF # not the default
@beta_blip_auto.sol
eof
Compulsory Keywords
Optional Keywords
3.9.6 Log-Likelihood Gain
MODE
MR_LLG combines the anisotropy correction and the likelihood function
(1) to calculate the log-likelihood gain for full
or partial molecular replacement solutions. Solutions are output to the
file FILEROOT.sol.
Example command script to rescore the solutions using a different resolution
range of data and a different spacegroup
beta_blip_llg.com
phaser ‹‹
eof
TITLe beta blip solution 6A P3121
MODE MR_LLG
HKLIn beta_blip.mtz
LABIn F=F SIGF = SIGF
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
ROOT beta_blip_llg # not the default
RESOlution 6.0
SPACegroup P 31 2 1
@beta_blip_auto.sol
eof
Compulsory Keywords
Optional Keywords
3.9.7 Packing
MODE
MR_PAK determines whether molecular replacement solutions pack in
the unit cell. Solutions that pack are output to the file FILEROOT.sol.
Example command script for determining whether a set of molecular replacement
solutions pack in the unit cell
beta_blip_pak.com
phaser ‹‹
eof
TITLe beta blip packing check
MODE MR_PAK
HKLIn beta_blip.mtz
LABIn F=F SIGF=SIGF
ENSEmble beta PDB beta.pdb IDENtity 100
ENSEmble blip PDB blip.pdb IDENtity 100
COMPosition PROTein MW 28853 NUM 1 #beta
COMPosition PROTein MW 17522 NUM 1 #blip
ROOT beta_blip_pak # not the default
PACK 1 # not the default
@beta_blip_auto.sol
eof
Compulsory Keywords
Optional Keywords
4. Experimental Phasing Modes
Phaser performs SAD phasing in two modes. In the Automated Experimental Phasing
mode, Phaser corrects for anisotropy, puts the data on absolute scale,
does a cell content analysis, refines heavy atom sites to optimize
phasing, and completes the model from log-likelihood gradient maps.
Alternatively, the SAD Phasing mode can be used,
which only refines heavy atom sites to optimize phasing, and completes
the model from log-likelihood gradient maps. For this mode, the data
should be pre-corrected for anisotropy and put on an absolute scale.
This mode should only be used as part of automation pipelines, where
the correct preparation of the data can be guaranteed and it saves cpu
time.
4.1 Automated Experimental Phasing
MODE
EP_AUTO combines the anisotropy correction, cell content analysis, and SAD Phasing modes
to automatically solve a structure by experimental phasing. The final solution
is output to the files FILEROOT.sol,
FILEROOT.mtz and FILEROOT.pdb. Many structures can be solved by running an automated experimental phasing job with
defaults.
Do SAD phasing of insulin. This is the minimum input, using all defaults (except the ROOT filename)
insulin_auto.com
phaser ‹‹
eof
MODE EP_AUTO
TITLe sad phasing of insulin with intrinsic sulphurs
HKLIn S-insulin.mtz
COMPosition NUCLeic SEQ S-insulin.seq
CRYStal insulin DATAset sad LABIn F+=F(+) SIG+=SIGF(+) F-=F(-) SIG-=SIGF(-)
LLGComplete CRYStal insulin COMPLETE ON SCATtering ELEMent S
ATOM CRYStal insulin PDB S-insulin_hyss.pdb
ROOT insulin_auto
eof
Compulsory Keywords
Optional Keywords
4.3 How to Define Atoms
Atom sites are defined with the ATOM
keyword. Atoms sites may be entered one at a time specifying fractional or orthogonal coordinates, occupancy and B-factor,
or from a PDB file, or from a mlphare-style HA file. The crystal to which the atoms correspond must be specified in the input.
4.4 How to Control Output
4.5. Basic Modes for Experimental Phasing
4.5.1 SAD Phasing
MODE
EP_SAD phases SAD data and completes the structure from log-likelihood gradient maps. The final solution
is output to the files FILEROOT.sol,
FILEROOT.mtz and FILEROOT.pdb
.
Do SAD phasing of insulin. This is the minimum input, using all defaults (except the ROOT filename)
insulin_sad.com
phaser ‹‹
eof
MODE EP_SAD
TITLe sad phasing insulin with intrinsic sulphurs
HKLIn S-insulin.mtz
CRYStal insulin DATAset sad LABIn F+=F(+) SIG+=SIGF(+) F-=F(-) SIG-=SIGF(-)
LLGComplete CRYStal insulin COMPLETE ON SCATtering ELEMent S
ATOM CRYStal insulin PDB S-insulin_hyss.pdb
ROOT insulin_sad
eof
Compulsory Keywords
Optional Keywords
Phaser can be controlled using keyword input. Not all keywords are relevant
for all modes of operation (the list of relevant keywords for each mode is given
with each mode above). Some keywords are only for single use, others have meaning
when used more than once. The input values of many parameters are constrained
to physically meaningful values. All non-compulsory parameters have defaults.
5.1 Preprocessor
Preprocessor commands may be used in the keyword
input to incorporate files, add comments or allow line continuation.
- @ filename
- To include a file in the input stream use the "@" (at) character
- Recursive application (can nest @ in files)
- # comment
- line of keyword input # comment
- All characters on a line after a "#" (hash) character are ignored
- line of keyword input &
line of keyword input continued
- Line continuation with the "&" (ampersand) character
- END GO RUN START STOP QUIT EXIT KILL
- End the input and start Phaser
- phaser HKLIN filename
- Simple keyword-value pairs may also be added to the command line. This enables the command
line to be used for input such as HKLIN, in traditional CCP4 style.
- KEYWord
- Courier font in blue means a keyword. Only the first four letters (sometimes
less) of any input keyword are required/recognized. The required characters
for each keyword are given in uppercase and those not required in lowercase.
Keywords are not case sensitive.
- <PARAMETER>
- Angle brackets mean a parameter value. Input strings are
case sensitive: the case of titles and filenames is preserved.
- { KEYWord
<X Y Z> }
- Curly brackets mean a group of keywords/parameters must come together.
- [ X | Y ]
- Square brackets and line separating options means X or Y.
- KEYWord <X Y Z>
- Italics mean the keyword/input is optional.
- *KEYWord
- Keywords marked with an asterix are for "expert" use only, or use in development.
- Constraint: X=%
- If 1<X<=100, stored value = X, else if 0<X<1, stored value = X*100, else error
- Default:
- Some default values are constants, others are set by Phaser after it has analyzed the input data.
5.3 Keywords
Most keywords only refer to a single parameters, and if used multiple times, the parameter will take last value input.
Some keywords are meaningful when entered multiple times. The order may or may not be important.
-
-
ATOM CRYStal <XTALID>
ELEMent <TYPE>
[ORTH|FRAC] <X Y Z>
OCCupancy <OCC>
[{ISOB <ISOB>} |
{[ANOU|USTAR] <HH KK LL HK HL KL>}]
FIXX FIXO FIXB BSWAP
{LABEL <SITE_NAME>}
- Definition
of atom position in crystal XTALID. B-factor defaults to isotropic and
Wilson B-factor if not set and ATOM BFACTOR WILSON is used (the
default). The B-factors can be set to another value with ATOM BFACTOR
VALUE
-
ATOM CRYStal <XTALID>
PDB <FILENAME>
- Definition of atom postiions in crystal XTALID using a pdb file.
-
ATOM CRYStal <XTALID>
HA <FILENAME>
- Definition of atom postiions in crystal XTALID using a ha file (from RANTAN, MLPHARE etc.).
-
ATOM BFACtor [ WILSON | {VALUE <B>} ]
- Reset all atomic B-factors to the Wilson B-factor or to B
Constraint: ISOB>=0
-
- *BINS
{MINimum
<L>} {MAXimum <H>}
{NUMber <N>} {WIDTh
<W>} {CUBIc <A B C>}
- The binning of the data. L = minimum number of bins, H = maximum number
of bins, N = number of bins, W = width of the bins in number of reflections,
A B C are the coefficients for the binning function A(S*S*S)+B(S*S)+CS where
S = (1/resolution). If N is given then the values of L and H are ignored.
Constraint: CUBIC coefficients restricted to monotonically increasing function: A >0, B >0, C >0 and either
(a) A=B=0 or (b) A=0 or (c) B=0
Default: BINS MIN 6 MAX 50 WIDTH 1000 CUBIC 0 1 0
-
- *BFACtor [WILSon|SPHEricity] [ON|OFF] SIGMA <SIGMA>
- Toggle
to set the Wilson restraint on the isotropic component of the atomic
B-factors, and the Sphericity restraint on the anisotropic B-factors,
and the sigma of the restraint, for experimental phasing.
Default: BFACTOR WILSON ON SIGMA 5
Default: BFACTOR SPHERICITY ON SIGMA 5
-
- *BOXSCALE <BS>
- Scale for box for calculating structure factors. The ensembles are put
in a box equal to (extent of molecule)*BS.
Constraint: BS >2.4
Default: BOXSCALE 4
-
- *CELL <A B C ALPHA BETA GAMMA>
- Unit cell dimensions
- *CELL CRYStal <XTALID> <A B C ALPHA BETA GAMMA>
- Unit cell dimensions for crystal (experimental phasing)
Constraint: A>0,B>0,C>0,ALPHA>0,BETA>0,GAMMA>0
Default: Cell read from MTZ file
-
- *CLMN
{SPHEre <SPHERE>}
{LMINimum <LMIN>}
{LMAXimum <LMAX>}
-
The radii or L values for the decomposition of the Patterson in
�ngstroms. Values depend on the geometric mean radius (GMR)of the
ENSEMBLE
Constraint: SPHERE>5, LMIN>0, LMAX>LMIN
Default: CLMN SPHERE <2*GMR> LMIN 2
-
- COMPosition [PROTein|NUCLeic]
[{MW <MW>}|{SEQuence <FILE>}]
NUMber <NUM>
- Composition of the crystals. The number of copies NUM of molecular
weight MW or SEQ given in fasta format (in a file FILE) of protein or nucleic
acid in the asymmetric unit.
Constraint: MW>0
Default: NUM=1
- COMPosition ATOM <ATOMTYPE>
NUMber <NUM>
- NUM atoms of ATOMTYPE are added to the composition.
- COMPosition ENSEmble <MODLID>
FRACtional <FRAC>
- Alternative way of defining composition. Fraction scattering is entered
explicitly for each ENSEMBLE.
Constraint: 0<FRAC<=1
-
- CRYStal <XTALID>
DATAset <WAVEID>
LABIn F=<F> SIGF=<SIGF>
- Columns of MTZ file to read for this (non-anomalous) dataset
- CRYStal <XTALID>
DATAset <WAVEID>
LABIn F(+)=<F+> SIGF(+)=<SIG+> F(-)=<F-> SIGF(-)=<SIG->
- Columns of MTZ file to read for this (anomalous) dataset
-
CRYStal <XTALID> DATAset <WAVEID>
SCATtering [
CUKA |
{WAVElength <L>}
{MEASured FP=<FP> FDP=<FDP>}
{ATOM <TYPE> FP=<FP> FDP=<FDP>} ]
- Set wavelength at which dataset was collected or set F-prime and F-double-prime explicitly.
Default: CRYSTAL <XTALID> SCATTERING CUKA
-
CRYStal <XTALID> DATAset <WAVEID>
FIXP FIXDP
- Fix Fp and Fdp for all scatterers in dataset
Default: Fp and Fdp refined
-
CRYStal <XTALID> DATAset <WAVEID>
RESOlution <HIRES> <LORES>
- Restrict resolution range of this dataset
- *CRYStal <XTALID> DATAset <WAVEID>
BINS {MINimum
<L>} {MAXimum <H>} {NUMber <N>} {WIDTh <W>} {CUBIc <A B C>}
- Binning parameters for this dataset
Default: As for BINS keyword
-
- EIGEn [ {READ <EIGENFILE>} | {WRITe [ON|OFF]} ]
- Read or write a file
containing the eigenvectors and eigenvalues. If reading, the eigenvalues
and eigenvectors of the atomic Hessian are read from the file generated
by a previous run, rather than calculated. This option must be used with
The job that generated the eigenfile
and the job reading the eigenfile must have identical (or default) input
for keyword NMAMethod. Use WRITe to control whether or not the eigenfile
is written when not using the READ mode.
Default: EIGEN WRITE ON
-
- ENSEmble <MODLID>
PDB <PDBFILE> [RMS|IDENtity] <NUM> {PDB
<PDBFILE> [RMS|IDENtity]
<NUM>}…
- The names of the PDB files used to build the ENSEMBLE, and either the expected RMS deviation of the coordinates to the
"real" structure or the percent sequence identity with the real sequence.
Constraint: NUM=% if IDENtity
- ENSEmble
<MODLID> HKLIn <MTZFILE>
F=<F> PHI=<P> EXTEnt
<EX EY EZ> RMS <RMS> CENTre
<CX CY CZ> PROTein MW <PMW> NUCLeic
MW <NMW>
- An ENSEmble defined from a map (via an mtz file). The molecular weight
of the object the map represents is required for scaling. The effective
RMS coordinate error is needed to judge how the map accuracy falls off with
resolution.. The extent is needed to determine reasonable rotation steps,
and the centre is needed to carry out a proper interpolation of the molecular
transform. The extent and the centre are both given in �ngstroms.
- *ENSEmble <MODLID>
BOXScale <BS>
- Scale for box for calculating structure factors. The ensemble is put
in a box equal to (extent of molecule)*BS.
Constraint: BS >2.4
Default: ENSEmble <MODLID> BOXSCALE 4
- *ENSEmble
<MODLID> BINS {MINimum
<L>} {MAXimum <H>} {NUMber
<N>} {WIDTh <W>} {CUBIc
<A B C>}
- Bins for the MODLID
Constraint: As for keyword BINS
Default: ENSEmble <MODLID> BINS MIN 6 MAX 200 WIDTH 1000 CUBIC 0 1 0
-
- *ENSIn <MODLID>
HKLIn <MTZFILE>
SCATtering <SCAT>
EXTEnt <EX EY EZ>
PR <P1 P2 P3 P4 P5 P6 P7 P8 P9>
PT <TX TY TZ>
- This
option can be used to read back a molecular transform computed in an
earlier Phaser job run in the MR_ENS mode.
May be useful if the spherical harmonic decomposition is very long.
This can only be used when repeating the search for a component
of the asymmetric unit with no (or the same) known fixed structure as
part of the search.
-
- *FFTS MINimum [ATOMS_MIN] MAXimum [ATOMS_MAX]
- The
minimum and maximum number of atoms of the range between which direct
summation and fft methods are tested to see which is faster for
structure factor and gradient calcuation (for this unit cell and
resolution). For a number of atoms below ATOMS_MIN direct structure
factor calculation is always used, and for a number of atoms above
ATOMS_MAX ffts are always used for the structure factor calculation and
the gradient calculations. Direct summation is always used for the
curvatures. Use FFTS MIN 0 MAX O to always use ffts.
Default: FFTS MIN 20 MAX 80
-
- FINAl [ROT|TRA] STEP [1|2|3] SELEct
[{SIGma <S>} |
{NUMber <N>} |
{PERCent <P>} |
ALL]
- Peaks satisfying selection criteria are saved. With neither ROT or TRA given, the criteria apply to both. With STEP [1|2|3]
not given, the criteria apply to all three.
Constraint: S>0,P=%
Default: FINAL SELECT PERCENT 75
- FINAL [ROT|TRA] CLUSter [ON|OFF]
{DUMP <NDUMP>}
{LOG [ON|OFF]}
- Toggle for CLUSTER selects clustered
peaks for saving. If clustered peaks are used, then NDUMP raw peaks will
be dumped to the output. If unclustered peaks are used
then you may still perform the clustering and log the results to the log
file (this may be time consuming).
Constraint: NDUMP>0
Default: FINAL SELECT PERCENT 75
Default: FINAL SELECT CLUSTER ON DUMP 20
-
- HAND [ON|OFF]
- Toggle for whether or not to use other hand of spacegroup in experimental phasing
Default: HAND OFF
-
- HKLIn <FILENAME>
- The mtz file containing the data
-
- HKLOut [ON|OFF]
- Flags for output of an mtz file containing the phasing information
Default: HKLOUT ON
-
- LABIn F = <F>
SIGF = <SIGF>
- Columns in mtz file. F must be given. SIGF should be given but is optional.
-
- LLGComplete NCYC <NMAX>
- Maximum
number of cycles of log-likelihood gradient structure completion. By
default, NMAX is 50, but this limit should never be reached, because
all features in the log-likelihood gradient maps should be assigned
well before 50 cycles are finished. This keyword should be used to
reduce the number of cycles to 1 or 2.
- LLGComplete CRYStal <XTALID>
{COMPlete [ON|OFF]}
{CLASh <clash>}
{SIGma <sigma>}
{SCATtering [ ELEMent <atomtype> { OR ELEMent <atomtype> }
|
REAL |
IMAG ]}
- Toggle
for structure completion by log-likelihood gradient maps, minimum
distance between atoms in log-likelihood gradient maps (default
determined by resolution), Z-score (sigma) for accepting peaks as new
atoms in log-likelihood gradient maps, and atom type(s) to be used for
log-likelihood gradient completion. If more than one element is entered
for log-likelihood gradient completion, the atom type that gives the
highest Z-score for each peak is selected. As an alternative to using
an atom type(s) for the calculation of the log-likelihood gradient map,
real or imaginary components of the map may be calculated.
Default: LLGCOMPLETE CRYSTAL <XTALID> COMPLETE OFF SCATTERING ELEMENT SE SIGMA 6
-
- MODE
[ ANO | CCA | NMA | MR_AUTO | MR_FRF | MR_FTF | MR_BRF | MR_BTF | MR_RNP | MR_LLG | MR_PAK | EP_AUTO | EP_SAD]
- The mode of operation of Phaser
-
-
NMAMethod
[RTB|CA|ALL]
{NRESidues <NRES>}
{MAXBlocks <MAXBLOCKS>}
{RADIus <RADIUS>}
{FORCe <FORCE>}
- Input for the normal mode analysis. Writes out pdb files perturbed along
normal mode(s). {RTB|CA|ALL} define the atoms used for the analysis.
RTB uses the rotation-translation block method, CA uses C-alpha atoms only
to determine the modes, and ALL uses all atoms to determine the modes (only
for use on very small molecules, less than 250 atoms). For the RTB analysis,
by default NRES is calculated so that it is as high as it can be without
reaching MAXBlocks.
The interaction radius used for the calculations and the force constant
can also be altered.
Default: NMAMETHOD RTB MAXBLOCKS 250 RADIUS 5 FORCE 1
-
- NMAPdb
{MODE <M1> {MODE
<M2>…}} {RMS <RMS>} {CLASh
<CLASH>} {STREtch
<STRETCH>} {MAXRms
<MAXRMS>} [FORWard|BACKward|TOFRo]
{DQ <DQ1> {DQ <DQ2>…}}
{COMBination }
- MODE keyword gives the mode along which to perturb the structure. If multiple modes are
entered, the structure is perturbed along all the modes AND combinations
of the modes given. There is no limit on the number of modes that can be
entered, but the number of pdb files explodes combinatorially. RMS
is the increment in rms �ngstroms between pdb files to be written. The structure
will be perturbed along each mode until either the C-alpha atoms clash with
(come within CLASH �ngstroms
of) other C-alpha atoms, the distances between C-alpha atoms STRETCH
too far (note that normal modes do not preserve the geometry) or the MAXRMS
deviation has been reached. The structure is perturbed either forwards or
backwards or to-and-fro (FORWARD|BACKWARD|TOFRO)
along the eigenvectors of the modes specified. Alternatively, the DQ
factors (as used by the Elnemo server (K. Suhre & Y-H. Sanejouand, NAR
2004 vol 32) ) by which to perturb the atoms along the eigenvectors can
be entered directly. The keyword COMBINATION
controls how many modes are present in any combination.
Default: NMAPDB MODE 7 RMS 0.3 TOFRO STRETCH 10.0 CLASH 2.0 MAXRMS 0.5 COMBINATION 3
-
- *MACANO
ANISotropic [ON|OFF] BINS
[ON|OFF] SOLK [ON|OFF] SOLB
[ON|OFF]
{NCYCle <NCYC>}
{MINImizer [BFGS|NEWTON|DESCENT]}
- Macrocycles for the refinement of SigmaN in the anisotropy correction
Default: MACANO ANIS ON BINS ON SOLK OFF SOLB OFF NCYC 50 MINIMIZER BFGS
- *MACANO OFF
- Turns off the anisotropy correction
-
- *MACMR ROT [ON|OFF]
TRA [ON|OFF]
{NCYCle <NCYC>}
{MINImizer [BFGS|NEWTON|DESCENT]}
- Molecular replacement refinement macrocycle. The macrocycles are performed in the order that they are entered
Default:
MACMR ROT ON TRA ON NCYC 20 MINIMIZER BFGS
-
- *MACSAD
K [ON|OFF]
B [ON|OFF]
SIGMA [ON|OFF]
XYZ [ON|OFF]
OCCupancy [ON|OFF]
BFACtor [ON|OFF]
FDP [ON|OFF]
SA [ON|OFF]
SB [ON|OFF]
SP [ON|OFF]
SD [ON|OFF]
{NCYCle <NCYC>}
{MINImizer [BFGS|NEWTON|DESCENT]}
{HIRES <HIRES>
{TARGet[NOT_ANOM_ONLY| ANOM_ONLY]}
- SAD refinement macrocycle. The macrocycles are performed in the order that
they are entered
Default:
MACSAD K OFF B OFF SIGMA OFF XYZ OFF OCC ON BFAC OFF FDP OFF SA OFF SB OFF SP OFF SD OFF NCYC 50 MINIMIZER BFGS
MACSAD K OFF B OFF SIGMA OFF XYZ ON OCC ON BFAC ON FDP OFF SA OFF SB OFF SP ON SD ON NCYC 50 MINIMIZER BFGS
-
- *MUTE [ON|OFF]
- Toggle
for running in "silent" mode, with no summary, logfile or verbose
output written to "standard output". Output can be extracted from
Results object in python, or from XML file.
Default: MUTE OFF
-
- *OUTLier [ON|OFF] <PROB>
- Control of the large unlikely E-value rejection. Outliers with a probability
less than PROB are rejected.
Default: OUTLIER ON 0.000001
-
- PACK <ALLOWED_CLASHES>
- NUMber of C-alpha atoms that clash within 2�ngstroms.
Default: PACK 0
-
- PERMutations [ON|OFF]
- Toggle for whether the order of the search set is to be permuted.
Default:
PERMUTATIONS OFF
-
- PURGe [ON|OFF] PERCent <PERC>
- Automated
Molecular Replacement only. Toggle for whether to purge the solution
list from the translation function and the refinement and phasing steps
(when searching multiple spacegroups) according to the best solution
found so far.
Default:
PURGE ON PERCENT 0.75
-
- RESCore [ROT|TRA] [ON|OFF]
- Toggle
for rescoring of fast search peaks. If neiter ROT or TRA are present,
toggle applies to both the rotation and translation function rescoring
(respectively), otherwise to the one specified.
Default: RESCore ON
-
- RESOlution <HIRES> <LORES>
- Resolution range in �ngstroms. If only one limit is given, it is the high
resolution limit; otherwise the limits can be in either order.
Constraint: HIRES>0,LORES>0
-
- ROOT <FILEROOT>
- Root filename for output files (e.g. FILEROOT.log)
Default: ROOT PHASER
-
- ROTAte
[ FULL |
{ROTAte AROUnd EULEr <A B G> RANGe <RANGE>} ]
- Sample all unique angles (FULL) or restrict the search to the region of +/- RANGE degrees around orientation given by EULER
Constraint: RANGE>0
Default: ROTAte FULL
-
- SAMPling [ROT|TRA] <SAMP>
- Sampling of search given in degrees for a rotation search and �ngstroms
for a translation search. Sampling for rotation search depends on the geometric mean radius (GMR) of the Ensemble and the
high resolution limit (dmin) of the search.
Constraint: SAMP>0
Default: SAMP = 2*atan(dmin/(4*GMR)) (MODE = MR_BRF)
Default: SAMP = 2*atan(dmin/(4*GMR)) (MODE = MR_FRF)
Default: SAMP = dmin/5; (MODE = MR_BTF)
Default: SAMP = dmin/3; (MODE = MR_FTF)
-
- *SCRIpt [ON|OFF]
- Write Phaser script file
Default: SCRIPT ON
-
- SEARch
ENSEmble <MODLID> {OR
ENSEmble <MODLID>}… {NUMber
<NUM>}
-
The ENSEMBLE
to be searched for in a rotation search or an automatic search. When multiple
ensembles are given using the OR keyword, the search is performed for each
ENSEMBLE
in turn. The final results are the best of all the searches (controlled
with the FINAL keyword). When the keyword is entered multiple times in the
MR mode, each SEARCH keyword refers to a new component of the structure.
If the component is present multiple times the sub-keyword NUMber can be
used (rather than entering the same SEARCH keyword NUMber times). If the
MR mode is being used with a fixed partial solution, only enter SEARCH keywords
(or associated NUMbers) for the components that remain to be found.
- SEARch MATThews [ON|OFF]
- The number of copies of the search components to search for is determined
by the Matthews coefficient. The stoichiometry of the search is given using
the NUM keyword above.
Default: SEARCH MATTHEWS OFF
-
- SGALternative
[ALL
| HAND | {TEST <SG>}]
- Alternative space groups to test in the translation function. All tests
all possible space groups, hand tests the given spacegroup and its enantiomorph
and <SG> tests the give space group.
Default: Only the SPACEGROUP is tested
-
- *SHANnon <SHARAT>
- Shannon sampling given by (2*SHARAT) for the Ensemble maps. Increase SHARAT to sharpen the sampling.
Constraint: SHARAT>1.1
Default: SHANNON 1.5
-
- SOLUtion
SET <ANNOTATION>
- Start new set of solutions
- SOLUtion
3DIM ENSEmble <MODLID> EULEr <A
B G> FIXR
- Rotation only solution. Use this keyword if only the orientation in 3
dimensions is known. This keyword is repeated for each case. A B G are the
Euler angles in degrees.
- SOLUtion
5DIM ENSEmble <MODLID> EULEr <A
B G> DEGEnerate [X|Y|Z] FRACtional
<U V> FIXR FIXT
- Use this keyword if the orientation in 3 dimensions and and the position
of the MODLID in only 2 dimensions is known. This keyword is repeated for
each case. A B G are the Euler angles in degrees. The keywords [X|Y|Z]specify
the degenerate direction and U V are the translation in the other two directions.
- SOLUtion
6DIM ENSEmble <MODLID> EULEr <A
B G> [ORTH|FRAC]
<X Y Z> FIXR FIXT
- This keyword is repeated for each known position and orientation of a
ENSEmble ID. A B G are the Euler angles and X Y Z are the translation.
- SOLUtion
TRIAl ENSEmble <MODLID> EULEr
<A B G> {DEGEnerate [X|Y|Z] FRACtional
<U V> } {SCORe <score>}
- Rotation List for translation function
-
- *SOLPARAMETERS <FSOL> <BSOL>
- Optionally change solvent parameters for Sigma(A) curves from the default
values. The results are not terribly sensitive to these parameters, which
affect only lower resolution data. FSOL and BSOL can be given in either
order, the lower number being taken as FSOL.
Constraint: 0<FSOL<1, BSOL>0
Default: SOLPARAMETERS FSOL 0.95 BSOL 300
-
- *SORT [ON|OFF]
- Sort the reflections into resolution order upon reading MTZ file, for performance gain in molecular replacement
Default: SORT ON
-
- SPACegroup
HALL <SG>
- Spacegroup may be altered from the one on the MTZ file to a spacegroup
in the same point group. The spacegroup name or number can be given e.g.
P 21 21 21 or 19. If the optional keyword HALL
is present, then the spacegroup is interpreted as a Hall symbol.
Default: Read from MTZ file
-
- *TARGet [LERF1|LERF2|CROWTHER]
- Target function for fast rotation searches (2)
Default: TARGet LERF1
- *TARGet [LETF1|LETF2|LETFL|LETFQ|CORRelation]
- Target function for fast translation searches (3)
Default: TARGet LETF1
-
- TITLe <TITLE>
- Title for job
Default: TITLE [no title given]
-
- TOPFiles <NUM>
- Number of top pdbfiles or mtzfiles to write to output.
Default: TOPFILES 1
-
- TRANslate
FULL
- Search volume for brute force translation function. Cheshire cell or
Primitive cell volume.
- TRANslate
LINE [ORTH|FRAC]
STARt <XS YS ZS> END
<XE YE ZE>
- Search volume for brute force translation function. Search along line.
- TRANslate
REGIon [ORTH|FRAC]
STARt <XS YS ZS> END
<XE YE ZE>
- Search volume for brute force translation function. Search region.
- TRANslate
AROUnd [ORTH|FRAC]
POINt <X Y Z> RANGe
<RANGE>
- Search volume for brute force translation function. Search within +/-
RANGE �ngstroms (not fractional coordinates, even if the search point
is given as fractional coordinates) of a point <X Y Z>.
- TRANslate
DEGEnerate [X|Y|Z]
- Search volume for brute force translation function. The search volume
is the plane perpendicular to the direction of the search.
Default: TRANSLATE FULL
-
- *VARSAD [
{K <N>}
{B <N>}
{SIGMA <N>}
{SA <B1 B2 …>}
{SB <B1 B2 …>}
{SP <B1 B2 …>}
{SD <B1 B2 …>}
- SAD variance parameters SA and SB (the real an imaginary components of Sigma Minus)
SP (Sigma Plus) and SD (Sigma Delta) by resolution bin and the overall scale (K) B-factor (B) and sigma-scale
(SIGMA) paramters.
-
- VERBose [ON|OFF] EXTRA
- Toggle to send verbose output to log file. If ON or OFF are not specified,
verbose is switched ON. If keyword EXTRA is present, then extra verbose information is logged.
Default: VERBOSE OFF
-
- XYZOut [ON|OFF]
- Toggle for output coordinate files.
Default: XYZOUT OFF (MODE MR_FRF, MODE MR_BRF)
Default: XYZOUT ON (other relevant modes)
6. XML Output (for developers)
Phaser can generate XML output from keyword input. XML output
should be used in preference to grepping logfiles when incorporating
Phaser into automation pipelines. Note that Phaser's python scripting ability is a more powerful way of calling Phaser for automation pipelines.
We would like to hear from developers who wish to incorporate Phaser into automation scripts using the XML functionality [email protected]
6.1 How to Use XML Output
6.1.1 Generating XML Output
Phaser outputs an XML file when called with the command line argument -xml followed by the filename for output.
phaser -xml <filename>
If no filename is given, Phaser exits immediately and writes an XML
file with filename PHASER.XML describing the error (type="FILE
OPENING", message="No XML filename").
6.1.2 Tags
All XML output is wrapped between "phaser" tags with the version
number for the Phaser executable and the operating system on which the
output was produced as attributes.
<phaser version="2.0" ostype="linux">
---All Phaser XML output---
<\phaser>
Names of files are output using the "file" tags with attributes
"type" specifying reflections ("HKL") or coordinates ("XYZ") and
attribute "format". Currently only "MTZ" and "PDB" formats are output.
<file type="HKL" format="MTZ">filename</file>
<file type="XYZ" format="PDB">filename</file>
Other XML tags are a combination of those suggested by the SPINE consortium and Phaser specific tags.
There is no schema. Please refer to the examples.
6.1.3 Error Handling
Successful Phaser execution (not necessarily structure solution!) is reported as
Failure during execution is reported as
More information about the type of error is given with
<error>
<name>ERROR_NAME</name>
<message>ERROR_MESSAGE</message>
</error>
Allowed values for ERROR_NAME are given in the table below, and specify the type of error.
The ERROR_MESSAGE gives more information as to the specific cause of the error.
| ERROR_NAME |
Failure due to … |
| SYNTAX |
Syntax error in keyword input |
| INPUT |
Input error (e.g. invalid value for an input parameter) |
| FILE OPENING |
Unable to open file (given in ERROR_MESSAGE) for reading or writing. |
| OUT OF MEMORY |
Memory exhaustion |
| FATAL RUNTIME |
Fatal runtime error (e.g. bug in Phaser) |
| UNHANDLED |
Other unhandled fatal error (e.g. bug in libraries) |
| UNKNOWN |
Other error (e.g. bug in compiler) |
6.1.4 Logfile Handling
Use the keyword MUTE ON
to prevent the writing of the logfile to standard output. Only the XML
file and other results files (mtz,pdb) will be produced by Phaser.
6.2 XML Output Examples
Below are example XML output files produced by running the most
popular modes of Phaser: anisotropy correction, automated molecular
replacement, cell content analysis, normal mode analayis, and SAD
phasing.
6.2.1 Anisotropy Correction
Output XML file for anisotropy correction of BETA-BLIP
<phaser version="2.0" ostype="linux">
<status>ok</status>
<anisotropy_info>
<file type="HKL" format="MTZ">beta_blip_ano.mtz</file>
<eigenB type="1">10.88</eigenB>
<eigenB type="2">10.88</eigenB>
<eigenB type="3">-21.76</eigenB>
<eigenB type="delta">32.64</eigenB>
<wilson type="K">20.25</wilson>
<wilson type="B">56.97</wilson>
</anisotropy_info>
</phaser>
6.2.2 Cell Content Analysis
Output XML file for Cell Content Analysis of BETA-BLIP
<phaser version="2.0" ostype="linux">
<status>ok</status>
<cell_content_analysis_info>
<mw>46375.000000</mw>
<solution id="0"</cca>
<Z>1</Z>
<probVM>0.995130</probVM>
<VM>2.340743</VM>
</solution>
</cell_content_analysis_info>
</phaser>
6.2.3 Normal Mode Analysis
Output XML file for Normal Mode Analysis of beta.pdb (modes 7 and 10, displacement forward only)
<phaser version="2.0" ostype="linux">
<status>ok</status>
<normal_modes id="1">
<file type="XYZ" format="PDB">beta_nma.1.pdb</file>
<displacement mode="7">0.000000</displacement>
<displacement mode="10">0.000000</displacement>
</normal_modes>
<normal_modes id="2">
<file type="XYZ" format="PDB">beta_nma.2.pdb</file>
<displacement mode="7">0.000000</displacement>
<displacement mode="10">47.306646</displacement>
</normal_modes>
<normal_modes id="3">
<file type="XYZ" format="PDB">beta_nma.3.pdb</file>
<displacement mode="7">46.881456</displacement>
<displacement mode="10">0.000000</displacement>
</normal_modes>
<normal_modes id="4">
<file type="XYZ" format="PDB">beta_nma.4.pdb</file>
<displacement mode="7">46.881456</displacement>
<displacement mode="10">47.306646</displacement>
</normal_modes>
<normal_modes id="5">
<file type="XYZ" format="PDB">beta_nma.5.pdb</file>
<displacement mode="7">93.762912</displacement>
<displacement mode="10">0.000000</displacement>
</normal_modes>
<normal_modes id="6">
<file type="XYZ" format="PDB">beta_nma.6.pdb</file>
<displacement mode="7">93.762912</displacement>
<displacement mode="10">47.306646</displacement>
</normal_modes>
</phaser>
6.2.4 Automated Molecular Replacement
Output XML file for Automated Molecular Replacement of BETA-BLIP
<phaser version="2.0" ostype="linux">
<status>ok</status>
<dataset_info>
<cell>
<a>75.11</a>
<b>75.11</b>
<c>133.31</c>
<alpha>90</alpha>
<beta>90</beta>
<gamma>120</gamma>
</cell>
<spacegroup>
<hall> P 32 2"</hall>
<number>154</number>
<lattice>P</lattice>
<operator id="0">32</operator>
<operator id="1">2</operator>
<operator id="2">1</operator>
</spacegroup>
<n_symops>6</n_symops>
<symmetry_operator id="0">
<aa>1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>1</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>1</cc>
<ctrans>0</ctrans>
</symmetry_operator>
<symmetry_operator id="1">
<aa>0</aa>
<ab>1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>-1</ba>
<bb>-1</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>1</cc>
<ctrans>0.666667</ctrans>
</symmetry_operator>
<symmetry_operator id="2">
<aa>-1</aa>
<ab>-1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>1</cc>
<ctrans>0.333333</ctrans>
</symmetry_operator>
<symmetry_operator id="3">
<aa>0</aa>
<ab>1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>-1</cc>
<ctrans>0</ctrans>
</symmetry_operator>
<symmetry_operator id="4">
<aa>-1</aa>
<ab>-1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>1</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>-1</cc>
<ctrans>0.666667</ctrans>
</symmetry_operator>
<symmetry_operator id="5">
<aa>1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>-1</ba>
<bb>-1</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>-1</cc>
<ctrans>0.333333</ctrans>
</symmetry_operator>
</dataset_info>
<solution id="1">
<file type="HKL" format="MTZ">beta_blip.1.mtz</file>
<file type="XYZ" format="PDB">beta_blip.1.pdb</file>
<llg>996.141543</llg>
</solution>
</phaser>
6.2.5 Automated Experimental Phasing
Output XML file for SAD phasing of insulin
<phaser version="2.0" ostype="linux">
<status>ok</status>
<dataset_info>
<cell>
<a>78.046</a>
<b>78.046</b>
<c>78.046</c>
<alpha>90</alpha>
<beta>90</beta>
<gamma>90</gamma>
</cell>
<spacegroup>
<hall> I 2b 2c 3</hall>
<number>199</number>
<lattice>I</lattice>
<operator id="0">21</operator>
<operator id="1">3</operator>
<operator id="2">3</operator>
</spacegroup>
<n_symops>24</n_symops>
<symmetry_operator id="0">
<aa>1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>1</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>1</cc>
<ctrans>0</ctrans>
</symmetry_operator>
<symmetry_operator id="1">
<aa>-1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>-1</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>0</cb> <cc>1</cc>
<ctrans>0</ctrans>
</symmetry_operator>
<symmetry_operator id="2">
<aa>1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>-1</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>0</cb> <cc>-1</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="3">
<aa>-1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>1</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>0</cb> <cc>-1</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="4">
<aa>0</aa>
<ab>1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>0</bb> <bc>1</bc>
<btrans>0</btrans>
<ca>1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>0</ctrans>
</symmetry_operator>
<symmetry_operator id="5">
<aa>0</aa>
<ab>-1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>0</bb> <bc>1</bc>
<btrans>0.5</btrans>
<ca>-1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>0</ctrans>
</symmetry_operator>
<symmetry_operator id="6">
<aa>0</aa>
<ab>-1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>0</bb> <bc>-1</bc>
<btrans>0</btrans>
<ca>1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="7">
<aa>0</aa>
<ab>1</ab> <ac>0</ac>
<atrans>0</atrans>
<ba>0</ba>
<bb>0</bb> <bc>-1</bc>
<btrans>0.5</btrans>
<ca>-1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="8">
<aa>0</aa>
<ab>0</ab> <ac>1</ac>
<atrans>0</atrans>
<ba>1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>1</cb> <cc>0</cc>
<ctrans>0</ctrans>
</symmetry_operator>
<symmetry_operator id="9">
<aa>0</aa>
<ab>0</ab> <ac>-1</ac>
<atrans>0</atrans>
<ba>1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>-1</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="10">
<aa>0</aa>
<ab>0</ab> <ac>-1</ac>
<atrans>0</atrans>
<ba>-1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>1</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="11">
<aa>0</aa>
<ab>0</ab> <ac>1</ac>
<atrans>0.5</atrans>
<ba>-1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0</btrans>
<ca>0</ca>
<cb>-1</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="12">
<aa>1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>1</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>0</cb> <cc>1</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="13">
<aa>-1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>-1</bb> <bc>0</bc>
<btrans>1</btrans>
<ca>0</ca>
<cb>0</cb> <cc>1</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="14">
<aa>1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>-1</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>0</cb> <cc>-1</cc>
<ctrans>1</ctrans>
</symmetry_operator>
<symmetry_operator id="15">
<aa>-1</aa>
<ab>0</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>1</bb> <bc>0</bc>
<btrans>1</btrans>
<ca>0</ca>
<cb>0</cb> <cc>-1</cc>
<ctrans>1</ctrans>
</symmetry_operator>
<symmetry_operator id="16">
<aa>0</aa>
<ab>1</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>0</bb> <bc>1</bc>
<btrans>0.5</btrans>
<ca>1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="17">
<aa>0</aa>
<ab>-1</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>0</bb> <bc>1</bc>
<btrans>1</btrans>
<ca>-1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="18">
<aa>0</aa>
<ab>-1</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>0</bb> <bc>-1</bc>
<btrans>0.5</btrans>
<ca>1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>1</ctrans>
</symmetry_operator>
<symmetry_operator id="19">
<aa>0</aa>
<ab>1</ab> <ac>0</ac>
<atrans>0.5</atrans>
<ba>0</ba>
<bb>0</bb> <bc>-1</bc>
<btrans>1</btrans>
<ca>-1</ca>
<cb>0</cb> <cc>0</cc>
<ctrans>1</ctrans>
</symmetry_operator>
<symmetry_operator id="20">
<aa>0</aa>
<ab>0</ab> <ac>1</ac>
<atrans>0.5</atrans>
<ba>1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>1</cb> <cc>0</cc>
<ctrans>0.5</ctrans>
</symmetry_operator>
<symmetry_operator id="21">
<aa>0</aa>
<ab>0</ab> <ac>-1</ac>
<atrans>0.5</atrans>
<ba>1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>-1</cb> <cc>0</cc>
<ctrans>1</ctrans>
</symmetry_operator>
<symmetry_operator id="22">
<aa>0</aa>
<ab>0</ab> <ac>-1</ac>
<atrans>0.5</atrans>
<ba>-1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>1</btrans>
<ca>0</ca>
<cb>1</cb> <cc>0</cc>
<ctrans>1</ctrans>
</symmetry_operator>
<symmetry_operator id="23">
<aa>0</aa>
<ab>0</ab> <ac>1</ac>
<atrans>1</atrans>
<ba>-1</ba>
<bb>0</bb> <bc>0</bc>
<btrans>0.5</btrans>
<ca>0</ca>
<cb>-1</cb> <cc>0</cc>
<ctrans>1</ctrans>
</symmetry_operator>
</dataset_info>
<solution_info>
<file type="XYZ" format="PDB">insulin.pdb</file>
<file type="HKL" format="MTZ">insulin.mtz</file>
<llg>81640.054723</llg>
</solution_info>
</phaser>
7. Python Scripting (for developers)
As an alternative to keyword input, Phaser can be called directly from a python
script. This is the way Phaser is called in Phenix and we encourage developers of
other automation pipelines to use the python scripting too.
In order to call Phaser in python you will need to have Phaser installed from source.
We would like to hear from developers who wish to incorporate
Phaser into automation scripts using the python scripting functionality
[email protected]
7.1 How to use Python scripting
7.1.1 Input-Objects, Run-Jobs, and Results-Objects
Using
Phaser through the python interface is similar to using Phaser through
the
keyword interface. Each mode of operation of Phaser described above is
controlled by an "input-object" (similar to the command script), has a
Phaser "run-job" which runs the Phaser executable for the corresponding
mode, and produces a
"result-object" (which includes the logfile text). The user input is
passed to the
"input-object" with a calls to set- or add- functions. Phaser is then
run with a call to the
"run-job" function, which takes the "input-object" for control. Results
are returned from the "result-object" with get-functions.
| Functionality |
Input-Object |
Run-Job |
Results-Object |
| Anisotropy Correction |
i = InputANO() |
r = runANO(i) |
ResultANO() |
| Cell Content Analysis |
i = InputCCA() |
r = runCCA(i) |
ResultCCA() |
| Normal Mode Analysis |
i = InputNMA() |
r = runNMA(i) |
ResultNMA() |
| Automated MR |
i = InputMR_AUTO() |
r = runMR_AUTO(i) |
ResultMR() |
| Fast Rotation Function |
i = InputMR_FRF() |
r = runMR_FRF(i) |
ResultMR_RF() |
| Brute Rotation Function |
i = InputMR_BRF() |
r = runMR_BRF(i) |
ResultMR_RF() |
| Fast Translation Function |
i = InputMR_FTF() |
r = runMR_FTF(i) |
ResultMR_TF() |
| Brute Translation Function |
i = InputMR_BTF() |
r = runMR_BTF(i) |
ResultMR_TF() |
| Refinement and Phasing |
i = InputMR_RNP() |
r = runMR_RNP(i) |
ResultMR() |
| Log-Likelihood Gain |
i = InputMR_LLG() |
r = runMR_LLG(i) |
ResultMR() |
| Packing |
i = InputMR_PAK() |
r = runMR_PAK(i) |
ResultMR() |
| Automated Experimental Phasing |
i = InputEP_AUTO() |
r = runEP_AUTO(i) |
ResultEP() |
| SAD Experimental Phasing |
i = InputEP_SAD() |
r = runEP_SAD(i) |
ResultEP() |
The major difference between running Phaser though the keyword
interface and running Phaser
though the python scripting is that the data reading and Phaser
functionality are separated. For the Phaser "run-job" functions, the
reflection data (for Miller indices, Fobs and SigmaFobs) are simply
arrays,
the spacegroup is given as a Hall string,
and the unitcell is given as an array of 6 numbers. This is an
important feature of the Phaser python scripting
as it means that the Phaser "run-job" functions are not tied to mtz
file input, but the data can be read in python from any file format,
and then the data passed to Phaser.
For the convenience of developers and users, the python scripting comes with data-reading jiffies to read data from mtz files.
(These are the same mtz reading jiffies that are used internally by Phaser when calling Phaser from keyword input.)
| Functionality |
Input-Object |
Run-Job |
Result-Object |
| Read Data for MR |
i = InputMR_DAT() |
r = runMR_DAT(i) |
ResultMR_DAT() |
| Read Data for EP |
i = InputEP_DAT() |
r = runEP_DAT(i) |
ResultEP_DAT() |
7.1.2. Input-Object set- and add-Functions
The syntax of the set- and add- functions on the "input-objects" mirror the keyword input.
Each "input-object" only has set- or add- functions corresponding to the keywords that are relevant for that mode.
Attempting to set a value on an "input-object" that is irrelevant for that mode will result in an error. This
differs from the keyword input, where the parser simply ignores any keywords that are not relevant to
the current mode. Some functions are common to all input-objects (described in the table below).
Note that
setting the spacegroup by name or number does not specify the setting. It is best to set the spacegroup
via the Hall symbol, which is unique to the full definition of the spacegroup.
| Input Objects |
Python Set Function |
| | ROOT filename |
i.setROOT(filename) |
| | MUTE [ON|OFF] |
i.setMUTE(True|False) |
| | TITLe title |
i.setTITL(title) |
| | VERBose [ON|OFF] |
i.setVERB(True|False) |
| | VERBose [ON|OFF] EXTRA |
i.setVERB_EXTRA(True|False) |
| * | SPACegroup name |
i.setSPAC_NAME(name) |
| * | SPACegroup number |
i.setSPAC_NUM(number) |
| * | SPACegroup Hall |
i.setSPAC_HALL(hall) |
| * | CELL a b c alpha beta gamm |
i.setCELL(a,b,c,alpha,beta,gamma) |
| * | Cell set from array of 6 numbers |
i.setCELL([a,b,c,alpha,beta,gamma]) |
| * except InputNMA |
7.1.3 Results-Object get-Functions
Data are extracted from the "result-objects" with get-functions.
The get-functions are mostly specific to the type of "result-object"
(described in sections below), but some are common to all
"result-objects" (described in table below).
Ralf Grosse-Kunstleve's scitbx::af::shared<double> array type is heavily used for passing of arrays into
the Phaser "input-objects" and extracting arrays from the Phaser "result-objects". This is a reference counted
array type that can be used directly in python and in C++. It is part of the Phaser installation, when Phaser
is installed from source. The scitbx (SCIentific ToolBoX) is part of the cctbx (Computational Crystallography
ToolBoX) which is hosted by sourceforge
| Results Objects |
Python Get Function |
| | Exit status "success" |
r.Success() |
| | Exit status "failure" |
r.Failure() |
| | Type of Error (see error table). SYNTAX errors are not thrown in python as they are generated by keyword input |
r.ErrorName() |
| | Message associated with error |
r.ErrorMessage() |
| | Text of Summary |
r.summary() |
| | Text of Logfile |
r.logfile() |
| | Text of Verbose Logfile |
r.verbose() |
| * | SpaceGroup Hall Symbol |
r.getSpaceGroupHall() |
| * | SpaceGroup Name (Hermann Mauguin, edited for CCP4 compatibility in R3 H3 R32 H32) |
r.getSpaceGroupName() |
| * | SpaceGroup Number |
r.getSpaceGroupNumber() |
| * | Number of symmetry operators |
r.getSpaceGroupNSYMM() |
| * | Number of primative symmetry operators |
r.getSpaceGroupNSYMP() |
| * | Symmetry operator #s, Rotation matrix element i,j (range 0-2) |
r.getSpaceGroupR(s,i,j) |
| * | Symmetry operator #s, Translation vector element i (range 0-2) |
r.getSpaceGroupT(s,i) |
| * | Unit Cell (array of 6 numbers) |
r.getUnitCell() |
| * except ResultNMA |
7.1.4 Error Handling
Exit status is indicated by Success() and Failure() functions of
the "result-objects". Success indicates successful execution of Phaser,
not that it has solved the structure! For molecular replacement jobs,
the foundSolutions() function indicates that Phaser has found one or
more potential solutions, the numSolutions() function returns how many
solutions were found and the uniqueSolution() function returns True if
only one solution was found. More detailed error information in the
case of Failure is given by ErrorName() and ErrorMessage().
Advanced Information: All errors are thrown and caught
internally by the "run-jobs", and so do not generate "Runtime Errors"
in the python script. In particular "INPUT" errors are not thrown by
the set- or add-functions of the "input-objects", but are stored in the
"input-object" and passed to the "result-object" once the "run-job" is
called. Results objects are derived from std::exception, and so can be
thrown. Function what() returns ErrorName() (not the ErrorMessage()).
7.1.5 Logfile Handling
Writing of the logfile to standard output can be silenced with the
i.setMUTE(True) function. The logfile or summary text can then be
printed to standard output with the print r.logfile() or print
r.summary() functions.
Advanced Information: Setting i.setMUTE(True) prevents real
time viewing of the progress of a Phaser job. This may present an
inconvenience for users. If you want to view the logfile information
but not have it go to standard output, Logfile text can be redirected
to a python string using an alternative call to the "run-job" function
that includes passing an "output-object" (which controls the Phaser
logging methods) on which the output stream has been set to a python
string. This feature of Phaser was developed thanks to Ralf
Grosse-Kunstleve.
beta_blip_logfile.py
from phaser import *
from cStringIO import StringIO
i = InputMR_DAT()
i.setHKLI("beta_blip.mtz")
i.setLABI("Fobs","Sigma")
i.setMUTE(True)
i = Output()
redirect_str = StringIO()
o.setPackagePhenix(file_object=redirect_str)
r = runMR_DAT(i,o)
7.2 Python Script Examples
Below are the detailed instructions for running the most popular
modes of Phaser: anisotropy correction, automated molecular
replacement, cell content analysis, normal mode analysis, and SAD
phasing.
Note that not all the functionality is available through the python
interface, particularly the "expert" functionality available through
the keyword interface, marked with an asterix in the keyword list.
7.1.1 Reading MTZ Files for Molecular Replacement
Input Object Type: InputMR_DAT
| Keyword Input |
Python Set/Add Function |
| HKLIn filename |
i.setHKLI(filename) |
| LABIn F=Fobs SIGF=Sigma |
i.setLABI(Fobs,Sigma) |
| RESOlution lim |
i.setHIRES(lim) |
| RESOlution lim1 lim2 |
i.setRESO(lim1,lim2 |
| SORT [ON|OFF] |
i.setSORT(True|False) |
Result Object Type: ResultMR_DAT
| Result |
Python Get Function |
| Miller Indices (array) |
r.getMiller() |
| F values (array) |
r.getF() |
| SIGF values (array) |
r.getSIGF() |
Example script for reading data from MTZ file beta_blip.mtz
Note that by default reflections are sorted into resolution order
upon reading, to achieve a performance gain in the molecular
replacement routines. If reflections are not being read from an MTZ
file with this script, reflections should be pre-sorted into resolution
order to achieve the same performance gain. Sorting is turned off with
the setSORT(False) function.
beta_blip_data.py
from phaser import *
i = InputMR_DAT()
HKLIN = "beta_blip.mtz"
F = "Fobs"
SIGF = "Sigma"
i.setHKLI(HKLIN)
i.setLABI(F,SIGF)
i.setMUTE(True)
r = runMR_DAT(i)
print r.logfile()
if r.Success():
hkl = r.getMiller()
fobs = r.getF()
sigma = r.getSIGF()
nrefl = min(10,hkl.size())
print "Data read from: " , HKLIN
print "Spacegroup Name (Hall symbol) = %s (%s)" % \
(r.getSpaceGroupName(), r.getSpaceGroupHall())
print "Unitcell = " , r.getUnitCell()
print "First ", nrefl , " reflections"
print "%4s %4s %4s %10s %10s" % ("H","K","L",F,SIGF)
for i in range(0,nrefl):
print "%4d %4d %4d %10.4f %10.4f" % \
(hkl[i][0],hkl[i][1],hkl[i][2],fobs[i],sigma[i])
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
7.2 Anisotropy Correction
Input Object Type: InputANO
| Keyword Input |
Python Set/Add Function |
| Read from MTZ file |
i.setREFL(HKL,F,SIGF) |
HKLIn filename
MTZ file to which output is appended
Data are not read from this file |
i.setHKLI(filename) |
LABIn F=Fobs SIGF=Sigma
Column label in output MTZ file Fobs_ISO and Sigma_ISO
Defaults F_ISO and SIGF_ISO |
i.setREFL_ID(Fobs,Sigma) |
| HKLOut [ON|OFF] |
i.setHKLO(True|False) |
| SORT [ON|OFF] |
i.setSORT(True|False) |
Result Object Type: ResultANO
| Result |
Python Get Function |
| Miller Indices (array) |
r.getMiller() |
| F values (array) |
r.getF() |
| SIGF values (array) |
r.getSIGF() |
| Corrected F (array) |
r.getCorrectedF() |
| Corrected SIGF (array) |
r.getCorrectedSIGF() |
| Correction Factor |
r.getCorrection() |
| Apply scale and correction factors to array |
new_array = r.getScaledCorrected(array) |
| Factor to put data on absolute scale |
r.WilsonK() |
| Wilson B factor |
r.WilsonB() |
| Measure of anisotropy |
r.getAnisoDeltaB() |
| Eigenvalues of anisotropy |
r.getEigenBs() |
| Eigenvectors and Eigenvalues or anisotropy |
r.getEigenSystem() |
| Name of output MTZ file |
r.getMtzFile() |
| Output MTZ file corrected F label |
r.getLaboutF() |
| Output MTZ file corrected SIGF label |
r.getLaboutSIGF() |
| Name of output XML file |
r.getXmlFile() |
Example script script for anisotropy correction of BETA-BLIP data
beta_blip_ano.py
from phaser import *
i = InputMR_DAT()
HKLIn = "beta_blip.mtz"
F = "Fobs"
SIGF = "Sigma"
i.setHKLI(HKLIn)
i.setLABI(F,SIGF)
i.setMUTE(True)
r = runMR_DAT(i)
if r.Success():
i = InputANO()
i.setSPAC_HALL(r.getSpaceGroupHall())
i.setCELL(r.getUnitCell())
i.setREFL(r.getMiller(),r.getFobs(),r.getSigFobs())
i.setREFL_ID(F,SIGF)
i.setHKLI(HKLIn)
i.setROOT("beta_blip_ano")
i.setMUTE(True)
del(r)
r = runANO(i)
if r.Success():
print "Anisotropy Correction"
print "Data read from: " , HKLIn
print "Data output to : " , r.getMtzFile()
print "Spacegroup Name (Hall symbol) = %s (%s)" % \
(r.getSpaceGroupName(), r.getSpaceGroupHall())
print "Unitcell = " , r.getUnitCell()
print "Principal components = " , r.getEigenBs()
print "Range of principal components = " , r.getAnisoDeltaB()
print "Wilson Scale = " , r.getWilsonK()
print "Wilson B-factor = " , r.getWilsonB()
hkl = r.getMiller();
f = r.getF()
sigf = r.getSIGF()
f_iso = r.getCorrectedF()
sigf_iso = r.getCorrectedSIGF()
corr = r.getCorrection()
nrefl = min(10,hkl.size())
print "First ", nrefl , " reflections"
print "%4s %4s %4s %10s %10s %10s %10s %10s" % \
("H","K","L",F,SIGF,r.getLaboutF(),r.getLaboutSIGF(),"Corr\'n")
for i in range(0,nrefl):
print "%4d %4d %4d %10.4f %10.4f %10.4f %10.4f %10.4f" % \
(hkl[i][0],hkl[i][1],hkl[i][2],f[i],sigf[i],f_iso[i],sigf_iso[i],corr[i])
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
7.2.3 Cell Content Analysis
Input Object Type: InputCCA
| Keyword Input |
Python Set/Add Function |
| COMPosition PROTein MW mw NUM num |
i.addCOMP_PROT_MW_NUM(mw,num) |
| COMPosition PROTein SEQ file NUM num |
i.addCOMP_PROT_FASTA_NUM(file,num)
where sequence in a fasta file
i.addCOMP_PROT_SEQ_NUM(sequence,num)
where sequence as a string, one letter code |
| COMPosition NUCLeic MW mw NUM num |
i.addCOMP_NUCL_MW_NUM(mw,num) |
| COMPosition NUCLeic SEQ seq NUM num |
i.addCOMP_NUCL_FASTA_NUM(file,num)
where sequence in a fasta file
i.addCOMP_NUCL_SEQ_NUM(sequence,num)
where sequence as a string, one letter code |
| COMPosition ATOM atomtype NUM num |
i.addCOMP_ATOM_NUM(atomtype,num) |
| COMPosition ENSEmble ens FRAC frac |
i.addCOMP_ENSE_FRAC(ens,frac) |
Result Object Type: ResultCCA
| Result |
Python Get Function |
| Molecular weight of the assembly used for VM calculations |
r.getAssemblyVM() |
| Number of multiples of the assembly within allowed VM range |
r.getNum() |
| Array of the multiples (Z) of the assembly within allowed VM range |
r.getZ() |
| Array of the values of VM corresponding to the multiples (Z) of the assembly |
r.getVM() |
| Array of the probabilities of VM corresponding to the multiples (Z) of the assembly |
r.getProb() |
| Most probable multiple (Z) of the assembly |
r.getBestZ() |
| VM of the most probable multiple (Z) of the assembly |
r.getBestVM() |
| Probability of the most probable multiple (Z) of the assembly |
r.getBestProb() |
| XML file name |
r.getXmlFile() |
| Optimal VM for spacegroup, unitcell and resolution |
r.getOptimalVM() |
| Optimal MW for spacegroup, unitcell and resolution |
r.getOptimalMW() |
Example script for cell content analysis of BETA-BLIP
beta_blip_cca.py
from phaser import *
i = InputMR_DAT()
HKLIN = "beta_blip.mtz"
F = "Fobs"
SIGF = "Sigma"
i.setHKLI(HKLIN)
i.setLABI(F,SIGF)
i.setMUTE(True)
r = runMR_DAT(i)
if r.Success():
if r.Success():
i = InputCCA()
i.setSPAC_HALL(r.getSpaceGroupHall())
i.setCELL(r.getUnitCell())
i.addCOMP_PROT_MW_NUM(28853,1)
i.addCOMP_PROT_MW_NUM(17522,1)
i.setMUTE(True)
del(r)
r = runCCA(i)
if r.Success():
print "Cell Content Analysis"
print "Molecular weight of assembly = " , r.getAssemblyMW()
print "Best Z value = " , r.getBestZ()
print "Best VM value = " , r.getBestVM()
print "Probability of Best VM = " , r.getBestProb()
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
7.2.4 Normal Mode Analysis
Input Object Type: InputNMA
| Keyword Input |
Python Set/Add Function |
| ENSEmble … |
As for InputMR_AUTO |
| NMAPdb MODE m1 MODE m2 … |
i.setNMAP_MODES([m1,m2…]) |
| NMAPdb RMS step |
i.setNMAP_RMS(step) |
| NMAPdb MAXRms distance |
i.setNMAP_MAXRMS(distance) |
| NMAPdb BACKward |
i.setNMAP_BACKWARD() |
| NMAPdb FORWard |
i.setNMAP_FORWARD() |
| NMAPdb TOFRo |
i.setNMAP_TO_AND_FRO() |
| NMAPdb COMBination nmax |
i.setNMAP_COMBINATION(nmax) |
| NMAPdb CLASh distance |
i.setNMAP_CLASH(distance) |
| NMAPdb STREtch distance |
i.setNMAP_STRETCH(distance) |
| NMAPdb DQ dq1 DQ dq2 … |
i.setNMAP_DQ([dq1,dq2…]) |
| NMAMethod RTB |
i.setNMAM_RTB() |
| NMAMethod CA |
i.setNMAM_CA() |
| NMAMethod ALL |
i.setNMAM_ALL() |
| NMAMethod NRESidues nres |
i.setNMAM_NRES(nres) |
| NMAMethod MAXBlocks maxblocks |
i.setNMAM_MAXBLOCKS(maxblocks) |
| NMAMethod RADIus radius |
i.setNMAM_RADIUS(radius) |
| NMAMethod FORCe force |
i.setNMAM_FORCE(force) |
| SCRIpt [ON|OFF] |
i.setSCRI(True|False) |
| XYZOut [ON|OFF] |
i.setXYZO(True|False) |
Result Object Type: ResultNMA
| Result |
Python Get Function |
| Number of total perturbations along combinations of normal modes |
r.getNum() |
| Array of all pdb files |
r.getPdbFiles() |
| Name of pdb file for perturbation #i |
r.getPdbFile(i) |
| Array of normal modes contributing to perturbation #i |
r.getModes(i) |
| Array of displacements along modes contributing to perturbation #i |
r.getDisplacements(i) |
| Script output file |
r.getSolFile() |
| Xml output file |
r.getXmlFile() |
Example script for normal mode analysis of BETA-BLIP. Note that the spacegroup and unitcell are not
required, and so the MTZ file does not need to be read to extract these parameters.
beta_nma.py
from phaser import *
i = InputNMA()
i.setROOT("beta_nma")
i.addENSE_PDB_ID("beta","beta.pdb",1.0)
i.setNMAP_MODES([7,10])
i.setNMAP_FORWARD()
i.setMUTE(True)
r = runNMA(i)
if r.Success():
print "Normal Mode Analysis"
for i in range(0,r.getNum()):
print "PDB file = ", r.getPdbFile(i)
displacement = r.getDisplacements(i)
mode = r.getModes(i)
for j in range(0,mode.size()):
print " Mode = " , mode[j], " Displacement = ", displacement[j]
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
7.2.5 Automated Molecular Replacement
Input Object Type: InputMR_AUTO
| Keyword Input |
Python Set/Add Function |
| Read from MTZ file |
i.setREFL(HKL,F,SIGF) |
| SGALternative num |
i.addSGAL_NUM(num) |
| SGALternative name |
i.addSGAL_NAME(name) |
| SGALternative HAND |
i.addSGAL_HAND(True|False) |
| SGALternative ALL |
i.addSGAL_ALL(True|False) |
| ENSEmble ens PDB pdbfile ID id |
i.addENSE_PDB_ID(ens,pdbfile,id) |
| ENSEmble ens PDB pdbfile RMS rms |
i.addENSE_PDB_RMS(ens,pdbfile,rms) |
| COMPosition … |
As for InputCCA |
| SEARch ENSEmble ens NUM num |
i.addSEAR_ENSE_NUM(ens,num) |
| SEARch ENSEmble ens1 {OR ENSEmble …} NUM num |
i.addSEAR_ENSE_OR_ENSE_NUM([ens1,…],num) |
| HKLOut [ON|OFF] |
i.setHKLO(True|False) |
| SCRIpt [ON|OFF] |
i.setSCRI(True|False) |
| XYZOut [ON|OFF] |
i.setXYZO(True|False) |
Result Object Type: ResultMR
| Result |
Python Get Function |
| Solutions were found (boolean) |
r.foundSolutions() |
| Number of Solutions that were found (int) |
r.numSolutions() |
| Only one solution found (boolean) |
r.uniqueSolution() |
| LLG values for all solutions in decreasing order |
r.getValues() |
| Script output file |
r.getSolFile() |
| Xml output file |
r.getXmlFile() |
| PDB files corresponding to solutions in decreasing LLG order |
r.getPdbFiles() |
| MTZ files corresponding to solutions in decreasing LLG order |
r.getMtzFiles() |
| LLG of top solution |
r.getTopValue() |
| PDB file of top solution |
r.getTopPdbFile() |
| MTZ file of top solution |
r.getTopMtzFile() |
List of details of solutions (rotation, translation)
in decreasing LLG order in "mr_solution" type |
r.getDotSol() |
Example script for automated structure solution of BETA-BLIP
beta_blip_auto.py
from phaser import *
i = InputMR_DAT()
i.setHKLI("beta_blip.mtz")
i.setLABI("Fobs","Sigma")
i.setHIRES(6.0)
i.setMUTE(True)
r = runMR_DAT(i)
if r.Success():
i = InputMR_AUTO()
i.setSPAC_HALL(r.getSpaceGroupHall())
i.setCELL(r.getUnitCell())
i.setREFL(r.getMiller(),r.getFobs(),r.getSigFobs())
i.setROOT("beta_blip_auto")
i.addENSE_PDB_ID("beta","beta.pdb",1.0)
i.addENSE_PDB_ID("blip","blip.pdb",1.0)
i.addCOMP_PROT_MW_NUM(28853,1)
i.addCOMP_PROT_MW_NUM(17522,1)
i.addSEAR_ENSE_NUM("beta",1)
i.addSEAR_ENSE_NUM("blip",1)
i.setMUTE(True)
del(r)
r = runMR_AUTO(i)
if r.Success():
if r.foundSolutions() :
print "Phaser has found MR solutions"
print "Top LLG = %f" % r.getTopValue()
print "Top PDB file = %s" % r.getTopPdbFile()
print "Spacegroup Name (Hall symbol) = %s (%s)" % \
(r.getSpaceGroupName(), r.getSpaceGroupHall())
else:
print "Phaser has not found any MR solutions"
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
7.2.6 Reading MTZ Files for Experimental Phasing
Input Object Type: InputEP_DAT
| Keyword Input |
Python Set/Add Function |
| HKLIn filename |
i.setHKLI(filename) |
| LABIn F=Fobs SIGF=Sigma |
i.setLABI(Fobs,Sigma) |
| RESOlution lim |
i.setHIRES(lim) |
| RESOlution lim1 lim2 |
i.setRESO(lim1,lim2) |
| CRYStal xtal DATAset wave LABIn F = F SIGF = SIGF
|
i.addCRYS_NORM_LABI(xtal,wave,F,SIGF) |
| CRYStal xtal DATAset wave LABIn F+ = Fp SIG+ = SIGp F- = Fn SIG- = SIGn
|
i.addCRYS_ANOM_LABI(xtal,wave,Fp,SIGp,Fn,SIGn) |
| CRYStal xtal DATAset wave RESOlution lim1 lim2 |
i.setCRYS_RESO(xtal,wave,lim1,lim2) |
Result Object Type: ResultEP_DAT
| Result |
Python Get Function |
| Miller Indices (array) |
r.getMiller() |
| Non-anomalous F values for crystal "xtal" and dataset "wave" (array) |
r.getF(xtal,wave) |
| Non-anomalous SIGF values for crystal "xtal" and dataset "wave" (array) |
r.getSIGF(xtal,wave) |
| Boolean flags for F (and SIGF) present for crystal "xtal" and dataset "wave" (array) |
r.getP(xtal,wave) |
| Anomalous F+ values for crystal "xtal" and dataset "wave" (array) |
r.getFpos(xtal,wave) |
| Anomalous SIGF+ values for crystal "xtal" and dataset "wave" (array) |
r.getSIGFpos(xtal,wave) |
| Boolean flags for F+ (and SIGF+) present for crystal "xtal" and dataset "wave" (array) |
r.getPpos(xtal,wave) |
| Anomalous F- values for crystal "xtal" and dataset "wave" (array) |
r.getFneg(xtal,wave) |
| Anomalous SIGF- values for crystal "xtal" and dataset "wave" (array) |
r.getSIGFneg(xtal,wave) |
| Boolean flags for F- (and SIGF-) present for crystal "xtal" and dataset "wave" (array) |
r.getPneg(xtal,wave) |
Below is a python script for reading SAD data from MTZ file S-insulin.mtz
insulin_data.py
from phaser import *
i = InputEP_DAT()
HKLIN = "S-insulin.mtz"
xtalid = "insulin"
waveid = "cuka"
i.setHKLI(HKLIN)
i.addCRYS_ANOM_LABI(xtalid,waveid,"F(+)","SIGF(+)","F(-)","SIGF(-)")
i.setMUTE(True)
r = runEP_DAT(i)
if r.Success():
hkl = r.getMiller()
Fpos = r.getFpos(xtalid,waveid)
Ppos = r.getPpos(xtalid,waveid)
Fneg = r.getFneg(xtalid,waveid)
Pneg = r.getPneg(xtalid,waveid)
print "Data read from: " , HKLIN
print "Spacegroup Name (Hall symbol) = %s (%s)" % \
(r.getSpaceGroupName(), r.getSpaceGroupHall())
print "Unitcell = " , r.getUnitCell()
nrefl = min(10,hkl.size())
print "First ", nrefl , " reflections with anomalous differences"
print "%4s %4s %4s %10s %10s %10s" % ("H","K","L","F(+)","F(-)","D")
i = 0
r = 0
while r < nrefl:
if Ppos[i] and Pneg[i] :
D = abs(Fpos[i]-Fneg[i])
if D > 0
print "%4d %4d %4d %10.4f %10.4f %10.4f" % \
(hkl[i][0],hkl[i][1],hkl[i][2],Fpos[i],Fneg[i],D)
r=r+1
i=i+1
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
7.2.7 Automated Experimental Phasing
Input Object Type: InputEP_AUTO
| Keyword Input |
Python Set/Add Function |
HKLIn filename
MTZ to which output data are appended
Data are not read from this file |
i.setHKLI(filename) |
| Input Data from array |
i.setCRYS_MILLER(miller) |
| Input Non-anomalous data for xtal/wave from array |
i.addCRYS_NORM_DATA(xtal,wave,F,SIGF,P) |
| Input Anomalous data for xtal/wave from array |
i.addCRYS_ANOM_DATA(xtal,wave,Fp,SIGp,Pp,Fn,SIGn,Pn) |
| CRYS xtal DATA wave SCAT CUKA |
i.setCRYS_SCAT_CUKA(xtal,wave) |
| CRYS xtal DATA wave SCAT WAVE lambda |
i.setCRYS_SCAT_WAVE(xtal,wave,lambda) |
| CRYS xtal DATA wave SCAT MEAS FP=fp FDP=fdp |
i.setCRYS_SCAT_MEAS(xtal,wave,fp,fdp) |
| CRYS xtal DATA wave SCAT ATOM type FP=fp FDP=fdp |
i.addCRYS_SCAT_ELEM(xtal,wave,type,fp,fdp) |
| CRYS xtal DATA wave SCAT FIXP FIXDP |
i.addCRYS_SCAT_FIX(xtal,wave,True,True) |
| LLGC CRYS xtal COMPLETE [ON|OFF] |
i.setLLGC_CRYS_COMPLETE(xtal,True|False) |
| LLGC CRYS xtal SCATTERING ELEMENT type |
i.addLLGC_CRYS_SCAT_ELEMENT(xtal,type) |
| LLGC CRYS xtal CLASH distance |
i.setLLGC_CRYS_CLASH(xtal,distance) |
| LLGC CRYS xtal SIGMA cutoff |
i.setLLGC_CRYS_SIGMA(xtal,cutoff) |
| ATOM CRYS xtal ELEM type FRAC x y z OCC occ |
i.addATOM(xtal,type,x,y,z,occ) |
| ATOM BFAC WILSON |
i.setATOM_BFACTOR_WILSON() |
| ATOM BFAC VALUE bfactor |
i.setATOM_BFACTOR_VALUE(bfactor) |
| ATOM CRYS xtal PDB pdbfile |
i.setATOM_PDB(xtal,pdbfile) |
| ATOM CRYS xtal HA hafile |
i.setATOM_HA(xtal,hafile) |
| HAND [ON|OFF] |
i.setHAND(True|False) |
| COMPosition … |
As for InputCCA |
Result Object Type: ResultEP
| Result |
Python Get Function |
| Log-likelihood of refined solution |
r.getLogLikelihood() |
| Miller Indices (array) |
r.getMiller() |
| Boolean array flaging reflections included in electron denisty |
r.getSelected() |
| Figures of merit for phased dataset (array) |
r.getFOM() |
| Amplitudes for weighted electrion density of phased dataset (array) |
r.getFWT() |
| Phases for weighted electrion density of phased dataset (array) |
r.getPHWT() |
| Phases for electrion density of phased dataset (array) |
r.getPHIB() |
| Amplitudes for log-likelihood gradient map |
r.getFLLG() |
| Phases for log-likelihood gradient map |
r.getPHLLG() |
| Atoms included in final solution for crystal xtal |
r.getAtoms(xtalid) |
| Atoms rejected from final solution for crystal xtal |
r.getRejectedAtoms(xtalid) |
| f' for atomtype "type" in crystal "xtald" dataset "wave" |
r.getFp(xtalid,wave,type) |
| f" for atomtype "type" in crystal "xtald" dataset "wave" |
r.getFdp(xtalid,wave,type) |
| Name of output MTZ file |
r.getMtzFile() |
| Name of output PDB file |
r.getPdbFile() |
| Name of output SOL file |
r.getSolFile() |
| Name of output XML file |
r.getXmlFile() |
There are also functions for extracting phasing statistics
| Result |
Python Get Function |
| Overall low resolution limit |
r.stats_lores() |
| Overall high resolution limit |
r.stats_hires() |
| Overall figure of merit for all reflections |
r.stats_fom() |
| Overall figure of merit for acentrics |
r.stats_acentric_fom() |
| Overall figure of merit for centrics |
r.stats_centric_fom() |
| Overall figure of merit for singleton |
r.stats_singleton_fom() |
| Overall number of reflections |
r.stats_num() |
| Overall number of acentric reflections |
r.stats_acentric_num() |
| Overall number of centric reflections |
r.stats_centric_num() |
| Overall number of singleton reflections |
r.stats_singleton_num() |
| Number of resolution bins for statistics |
r.stats_numbins() |
| Binwise low resolution limit |
r.stats_by_bin_lores(bin_number) |
| Binwise high resolution limit |
r.stats_by_bin_hires(bin_number) |
| Binwise middle of resolution range |
r.stats_by_bin_midres(bin_number) |
| Binwise figure of merit for all reflections |
r.stats_by_bin_fom(bin_number) |
| Binwise figure of merit for acentrics |
r.stats_by_bin_acentric_fom(bin_number) |
| Binwise figure of merit for centrics |
r.stats_by_bin_centric_fom(bin_number) |
| Binwise figure of merit for singleton |
r.stats_by_bin_singleton_fom(bin_number) |
| Binwise number of reflections |
r.stats_by_bin_num(bin_number) |
| Binwise number of acentric reflections |
r.stats_by_bin_acentric_num(bin_number) |
| Binwise number of centric reflections |
r.stats_by_bin_centric_num(bin_number) |
| Binwise number of singleton reflections |
r.stats_by_bin_singleton_num(bin_number) |
Example script for SAD phasing for insulin
insulin_sad.py
from phaser import *
from cctbx import xray
i = InputEP_DAT()
HKLIN = "S-insulin.mtz"
xtalid = "insulin"
waveid = "cuka"
i.setHKLI(HKLIN)
i.addCRYS_ANOM_LABI(xtalid,waveid,"F(+)","SIGF(+)","F(-)","SIGF(-)")
i.setMUTE(True)
r = runEP_DAT(i)
if r.Success():
hkl = r.getMiller()
Fpos = r.getFpos(xtalid,waveid)
Spos = r.getSIGFpos(xtalid,waveid)
Ppos = r.getPpos(xtalid,waveid)
Fneg = r.getFneg(xtalid,waveid)
Sneg = r.getSIGFneg(xtalid,waveid)
Pneg = r.getPneg(xtalid,waveid)
i = InputEP_AUTO()
i.setSPAC_HALL(r.getSpaceGroupHall())
i.setCELL(r.getUnitCell())
i.setCRYS_MILLER(hkl)
i.addCRYS_ANOM_DATA(xtalid,waveid,Fpos,Spos,Ppos,Fneg,Sneg,Pneg)
i.setATOM_PDB(xtalid,"S-insulin_hyss.pdb")
i.setLLGC_CRYS_COMPLETE(xtalid,True)
i.addLLGC_CRYS_SCAT_ELEMENT(xtalid,"S")
i.addCOMP_PROT_FASTA_NUM("S-insulin.seq",1.)
i.setHKLO(False)
i.setSCRI(False)
i.setXYZO(False)
i.setMUTE(True)
r = runEP_AUTO(i)
if r.Success():
print "SAD phasing"
print "Data read from: " , HKLIN
print "Data output to : " , r.getMtzFile()
print "Spacegroup Name (Hall symbol) = %s (%s)" % \
(r.getSpaceGroupName(), r.getSpaceGroupHall())
print "Unitcell = " , r.getUnitCell()
print "LogLikelihood = " , r.getLogLikelihood()
atom = r.getAtoms(xtalid)
print atom.size(), " refined atoms"
print "%5s %10s %10s %10s %10s %10s" % \
("atom","x","y","z","occupancy","u-iso")
for i in range(0,atom.size()):
print "%5s %10.4f %10.4f %10.4f %10.4f %10.4f" % \
(atom[i].scattering_type,atom[i].site[0],atom[i].site[1],atom[i].site[2],atom[i].occupancy,atom[i].u_iso)
hkl = r.getMiller();
fwt = r.getFWT()
phwt = r.getPHWT()
fom = r.getFOM()
nrefl = min(10,hkl.size())
print "First ", nrefl , " reflections"
print "%4s %4s %4s %10s %10s %10s" % \
("H","K","L","FWT","PHWT","FOM")
for i in range(0,nrefl):
print "%4d %4d %4d %10.4f %10.4f %10.4f" % \
(hkl[i][0],hkl[i][1],hkl[i][2],fwt[i],phwt[i],fom[i])
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
else:
print "Job exit status FAILURE"
print r.ErrorName(), "ERROR :", r.ErrorMessage()
8. Version History
- Phaser–2.0
- First release of experimental phasing (SAD only)
- Phaser–1.3.1
- Minor bug fixes after Phenix-1.1a release
- R-factor reported by MR_RNP and MR_LLG modes (verbose output)
- Phaser–1.3
- Released as part of Phenix-1.1a
- New Normal Mode Analysis of structures
- New Cell Content Analysis mode
- Improved Automated MR
- Map coefficients appended to input mtz file
- Score stored in .sol file is Z-score rather than LLG
- Final phasing and refinement to full resolution on mtz file
- Packing of RNA/DNA
- Packing to form close packed oligomeric complexes
- Waters in pdb files excluded from packing
- Only the most homologous model in an ensemble is used for packing analysis
- Reduced memory requirements
- Composition/MW determined from sequence
- Automated MR now available as a python script
- Bug fixes for Phaser–1.2
- Phaser–1.2
- First "official" release of Phaser software
- Automated solution of structures
- New Likelihood Enhanced fast Translation Function (LETF)
- Ability to use electron density maps as molecular replacement models
- Rigid body solution refinement against maximum likelihood target
- Pruning of duplicate solutions from solution list
- Searching of multiple alternative space groups
- Searching of multiple alternative models
- Better minimizers
- Phaser–1.1
- Bug in anisotropy refinement corrected
- Phaser–1.0
9. References
- Read, R.J. (2001). Pushing
the boundaries of molecular replacement with maximum likelihood. Acta
Cryst. D57, 1373-1382
- Storoni, L.C., McCoy, A.J. & Read, R.J. (2004).
Likelihood-enhanced
fast rotation functions. Acta Cryst D60, 432-438
- McCoy, A.J., Grosse-Kunstleve, R.W., Storoni,
L.C. & Read, R.J. (2005). Likelihood-enhanced
fast translation functions. Acta Cryst D61, 458-464
- McCoy, A.J., Storoni, L.C. and Read, R.J. (2004) Simple
algorithm for a maximum-likelihood SAD function Acta Cryst. D60, 1220-1228.






