| Python-based Hierarchical ENvironment for Integrated Xtallography |
| Documentation Home |
MRage: automated molecular replacement
MRage is generic molecular replacement program that is designed to run efficiently with a large number of possible models on parallel hardware. It accepts multiple model definitions that are internally processed with integrated tools to search models and used for molecular replacement. Identified solutions can be used to speed up the search even further by model assembly algorithms and limiting search space. InputThe composition of the unit cell is specified by inputting components, and potential models for the corresponding component in a hierarchical way. It is recommended to specify the sequence of a component, which allows for better handling of partial models (see Composition stage). A stoichiometry of each component in the final assembly is also assigned at this stage. The overall count of assemblies in the unit cell can either be input or determined automatically from the Matthews coefficient. In case no model is known for a component, the list of potential models should be left empty. ModelsModels for a component can be specified stepwise with the amount of preprocessing required. The following options are available:
These steps are performed on-demand, i.e. if no clear solution could be found with available ensembles, the program processes the next template, or fetches the next homology search hit, etc. Symmetry explorationSpecifies the set of space groups to try in the search. Currently, there are three options available:
Space group exploration is exhaustive and can take several macrocycles to complete. Space groups with clearly inferior results to others are incrementally removed from the search until the correct space group is established. Search organizationMRage employs a sequential search algorithm, in which molecules are found consecutively. The algorithm consists of several stages, and also allows the exploration of potential space groups. Composition stageThe composition of each partial structure (located in previous macrocycles) is compared with the composition of the unit cell. Available models that fit in the missing composition are selected for search. In case the modelled sequence for a search model is known, the program can account for the fact that a model may only be a partial model for a component (e.g. one domain only). Search stageCalculations are organized into a search tree that is explored according to depth-first traversal. For the underlying calculations, available functions from phaser [PHASER] are used. For each partial structure and applicable model, a rotation function is calculated. This is then followed up by a translation function calculation. Each translation function peaks are checked for packing clashes, and if accepted, checked whether are significant, probable solutions or not. Calculations are performed in the order of peak score (RF/TF), model quality (sequence identity) or partial structure quality (LLG score). Exploration continues until a significant solution is found (quick mode) or all possibilities has been exhausted (full mode). There is also no thresholding step for rotation (rationale: weak signal in rotation function) and translation function peaks (rationale: packing check fast). As a consequence, more work is performed, but simultaneously this allows weighting translation function results with packing overlap, and also avoids pipe stalls, which would adversely effect scaling in a parallel environment. In quick mode, if a significant solution is found, conventional search terminates. However, all models that are alternatives of the model that gave this clear solution are superposed onto the solution and refined. This allows the quick evaluation of model quality for a potentially large number of alternative models. Currently, SSM [SSM] is used for superposition. Parallelisation is done at a function level. Calculations are dispatched until the number of assigned CPUs are filled up. Execution can either take place in a shared memory environment using threads and processes or using a submission queue (currently SGE, LSF and PBS are supported). The degree of parallelisation scales with the complexity of the search, and given unlimited resources, reduces the runtime to a constant. However, in simple cases (e.g. a good model that gives a clear rotation and translation peak), no parallel resources can be used. RefinementPeaks above a certain threshold of the best peak and those that are identified as significant solutions are subjected to refinement. After refinement is completed, a second thresholding step is performed. Top peaks will be subjected to post-processing and also carried forward for the next macrocycle. Significant peaks that are below the threshold (e.g. a correct solution for a small domain) are also input to the post-processing step, but will not be propagated to the next cycle. Post-processingSignificant solutions are analysed for specific features so that the search could be improved.
No actual calculation is performed, only the possibilities are noted. In case the given solution enters the next Search stage (as determined by Composition stage), these possibilities will be scored as a priority. Space group explorationSearch progress in possible space groups is compared. Space groups having significant solutions or above the threshold (determined by the best score over all space groups) are propagated to the next macrocycle. OutputThe program logs all operations it performs. However, this may be difficult to follow on the screen, especially when multiple space groups are explored, and therefore, after each macrocycle, a summary of all searches performed is printed. Models generated during the solution process are also saved. This includes structure files fetched from the PDB, but also alignments and homology searches performed. SolutionsIf the search finishes, solutions are written out in all likely space groups. For each space group, the top solution is selected, and all partial solutions above a threshold relative to the top solution is considered. Solutions that are fully "contained" in a higher scoring solution (i.e. all models correspond to a model in the higher scoring solution) are discarded. This way, the top solution is output even if the composition is overestimated, but at the same time removing incomplete solutions (e.g. 14 molecules out of the total 15) that cannot be distinguised based on the final score. Solutions are written out in an internal compressed format, which is capable of storing a high number of solutions without onerous storage requirements, and can be manipulated with provided tools. UsageGraphical interfaceAs with the generic Phaser GUI, two interfaces are available for running MRage, one designed for single-component searches with minimal customization of input models, and an advanced interface that allows all available features to be used. These are launched from a single button in the main GUI, which will initially pop up an information dialog describing the available options.  The simple GUI is centered around a single input control allowing any combination of files to be dragged in:
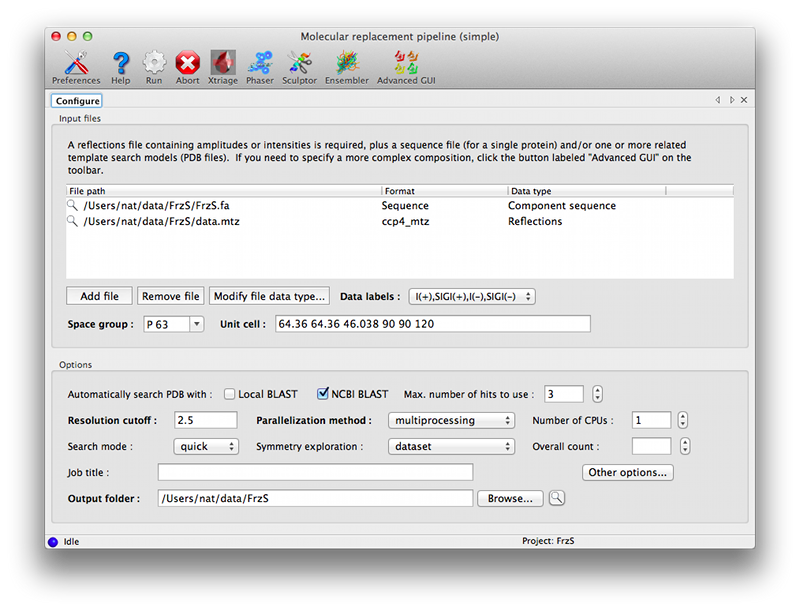 In the simplest cases, only the data and sequence are required; you should then select a homology search method in the Options section. "NCBI BLAST" requires internet access; "Local BLAST" requires that BLAST is installed and available on the command line. At this point you can run the program without further input.
Advanced GUIThis version of the GUI moves the description of the crystal contents and search information to a separate tab, with input options grouped by component. The first tab is essentially similar to the simple GUI, but the only input field is for the reflections file. 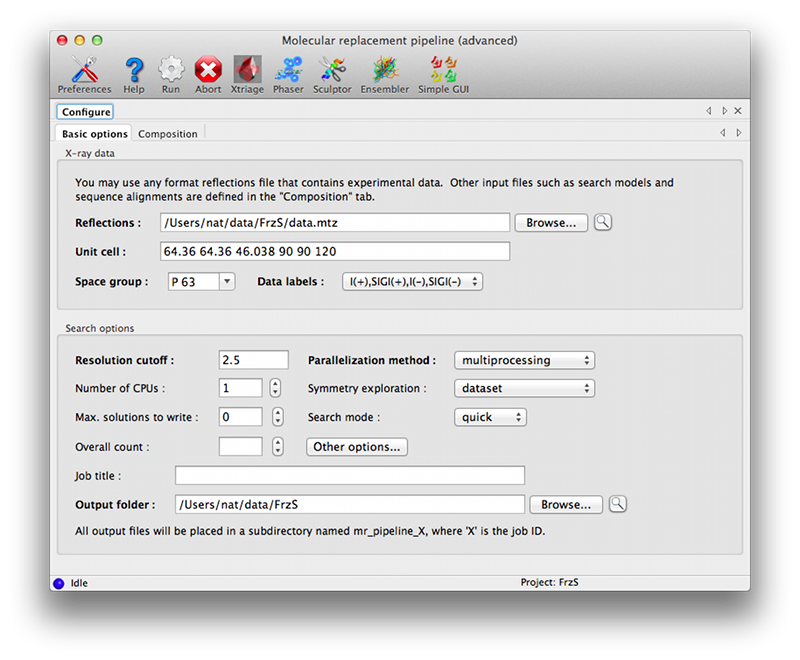 The composition tab contains another set of tabs for each unique component. These allow the full range of input files to be specified; some of these have additional parameters (such as the mandatory sequence identity for ensembles and model collections). The "Basic info" tab allows the NCBI BLAST search to be run interactively if desired (the results will be loaded as a homology result). A link to the HHPred server, which does not allow automated queries, is available on the "Homology searches" tab. 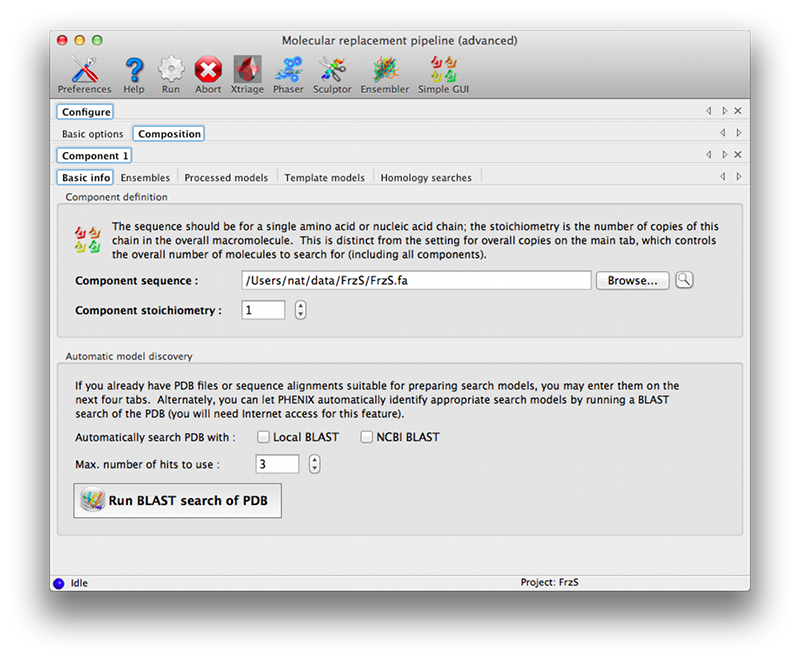 You must specify the component stoichiometry, i.e. the number of copies of the component in the overall assembly. This is distinct from the "Overall count" parameter in the main tab: for instance, if you are trying to solve a structure of a hexamer bound to a monomer and expect two copies of the complex in the asymmetric unit of the crystal, the overall count is two, and the stoichiometry of the components is 6 and 1 for the hexamer and monomer respectively. Command linephaser.MRage [command-line switches] [PHIL-format parameter files] -h, --help show this help message and exit --show-defaults print PHIL and exit -i, --stdin read PHIL from stdin as well -v, --verbosity set verbosity level (DEBUG,INFO,WARNING,VERBOSE) Everything not starting with a dash('-') is interpreted as a PHIL argument. This can be a PHIL-format file containing parameters or a command-line assignment. References
List of all MRage keywords
-------------------------------------------------------------------------------
Legend: black bold - scope names
black - parameter names
red - parameter values
blue - parameter help
blue bold - scope help
Parameter values:
* means selected parameter (where multiple choices are available)
False is No
True is Yes
None means not provided, not predefined, or left up to the program
"%3d" is a Python style formatting descriptor
-------------------------------------------------------------------------------
hklin= None
labin= None
resolution_cutoff= 2.5
mode= *quick full
symmetry_exploration= pointgroup enantiomorph *dataset
packing_pool= 20
rotation_peaks_cutoff= 0.75
post_refinement_cutoff= 0.75
final_selection_cutoff= 0.75
b_factor_refinement= True
significant_peak_threshold= 7.0
sculptor_protocols= 11 10 13 12 1 3 2 5 4 7 6 9 8 *all minimal
template_equivalence= False
assembly_acceptance_policy= observed *always never
assembly_ensemble_creation_policy= *observed always never
exclude_pdb_ids= None Developer option to prevent specific structures from
being used as search models, in order to test performance
with lower sequence identity structures.
crystal_symmetry
unit_cell= None
space_group= None
output
root= "mrage"
max_solutions_to_write= 0
gui_base_dir= None Base directory in which the PHENIX GUI will create a
folder containing the results. Ignored when running the
program from the command line.
save_aniso_data= False
job_title= None Job title in PHENIX GUI, not used on command line
composition
count= None Overall number of copies of the complete molecule (which may be
multiple components). If left blank, this will be guessed based on
the expected solvent content.
component
sequence= None
stoichiometry= 1 Number of copies of this component relative to the
overal macromolecule. This is separate from the overall
count parameter in the Basic Options tab, which defines
the number of copies of the complete unit. For instance,
if you expect the basic molecule to be two copies of this
component and one copy of a second component, the
stoichiometry is 2. If you are searching for an unknown
number of copies of a single component, you should leave
the stoichiometry set to 1.
ensemble
coordinates
pdb= None
identity= None
error_translation= *default floored_chothia_and_lesk rmsd Identity
to RMS conversion
model_collection
trim= False
coordinates
pdb= None
identity= None
error_translation= *default floored_chothia_and_lesk rmsd Identity
to RMS conversion
template
pdb= None
alignment= None
homology
file_name= None
max_hits= 3
search
services= local ncbi Performs a BLAST search of the PDB using either
a locally installed version of BLAST and the PDB sequence
database (requires separate installation), or the NCBI's
BLAST server (requires Internet access).
max_hits= 3
assembly Define known assembly
use_assembled= False Use assembly as model in search or only for completion
local_search Define local search extent
sweep= 5 Rotational search radius
extent= 5 Translational search extent
component Define a component of the assembly
file_name= None Assembly component structure
operation Model transformation to participate in the assembly
euler= 0 0 0 Rotation
displacement= 0 0 0 Translation
queue
technology= lsf threading sge *multiprocessing pbs
cpus= 1
submission_command= None
qslot_cpus= 1
simple_run Parameters for simplified GUI - not intended for command-line use.
enable= False GUI parameter - not intended for command-line use.
component_sequence= None
template_model= None
alignment= None
blast_services= local ncbi Performs a BLAST search of the PDB using either
a locally installed version of BLAST and the PDB sequence
database (requires separate installation), or the NCBI's
BLAST server (requires Internet access).
max_blast_hits= 3
| ||||