Contents
- Programming: Nigel W. Moriarty, Dorothee Liebschner, Nat Echols, Billy K. Poon, Pavel Afonine, Jeff Headd (LBL)
- Concept: Herb Klei (BMS)
The structure comparison GUI is a tool for parallel validation and analysis of near-identical protein structures, such as different crystal forms, various mutants, or NCS-related copies. It displays validation outliers and regions of difference for a set of individual chains on a spreadsheet-like grid, which is linked to the graphics windows (Coot and PyMOL). If desired, both the extracted chains and electron density maps can be superimposed into a common frame of reference. These chains may subsequently be edited in Coot to ensure consistency and/or fix errors, and recovered in their original orientations for further refinement and rebuilding.
We have included a set of representative structures of human Protein Kinase A (PKA) for comparison in the Phenix distribution. To automatically set up a new project with these data, click "New Project" in the main GUI toolbar and then "Set up tutorial data..." in the dialog which appears, and select "Protein Kinase A (Structure Comparison)" from the drop-down menu.
- Only protein chains are supported at this time. RNA/DNA are not supported.
- Only X-ray diffraction data are supported. It is planned to extend usage for cryo-EM and neutron data in the future.
- Only a single set of chains can be compared at a time; if you have a set of heterogeneous complexes, the components need to be dealt with separately.
- This tool is of limited use for comparing proteins which diverge significantly in sequence; the program will quit with a warning if the fractional identity drops below 0.8 (this can be adjusted if desired). See below for additional details.
The tutorial video is available on the Phenix YouTube channel and covers the following topics:
At a minimum, two or more PDB files containing near-identical chains are needed. A sequence file containing a single amino-acid sequence can be used to define the chain to be compared. It is recommended to also provide either X-ray data or pre-calculated map coefficients (the latter are preferred since these calculations are the slowest part of the program). The files generated by phenix.refine or phenix.maps are suitable for the latter. Each PDB file may contain an unlimited number of matching chains; in the PKA example, the structure 1syk has two copies of the catalytic subunit, while the rest contain only one. Non-matching chains will be ignored.
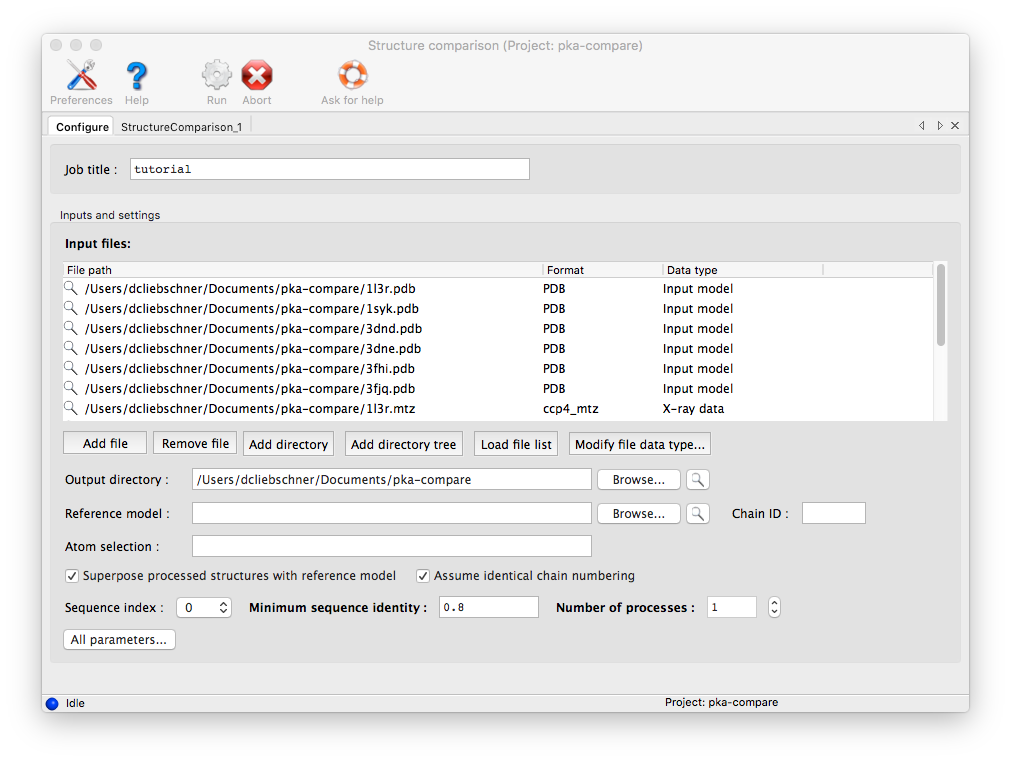
There are several options to open files or file pairs:
Files may be dragged into the "Input files" window in the center of the Structure Comparison GUI.
Files can be opened one by one by using the "Add File" button.
If you have a directory containing multiple allowed file types, you may simply drag the directory in, and it will automatically guess the appropriate use for each file. If the PDB and reflection files use similar root filenames, or occur in pairs in the same directories, the association between them will be guessed automatically. For instance, the following pair of files will automatically be recognized as model and experimental data:
1syk.pdb 1syk.mtz
while these will be recognized as model and map coefficients:
1syk.pdb 1syk_map_coeffs.mtz
Similarly, the "Add Directory" button can be used to open a directory and the program tries to automatically guess the file pairs.
"Add Directory Tree" searches for file pairs in a directory tree.
The map calculations have been parallelized, and this part of the program scales extremely well over multiple processors. The GUI will automatically be set to use ncpu=1 processes, but you may increase this if desired.
The most important configurable option is the choice of superposing the models and maps prior for viewing together in Coot or PyMOL. This is very advantageous for comparing regions of difference, instead of switching between views for each chain. However, it has the drawback of removing the chains being analyzed from their original context, possibly obscuring other features that account for the observed structures. If you choose to turn on the superpositioning, the extracted chains will be written out as separate files in the results directory (along with reoriented maps for each chain, if available). Otherwise, the original models will be used for viewing. (Note that the analyses which depend on local environment in the crystal will still be run on the original models, and the coordinates mapped to their new positions.)
In addition to the models actually being compared, you may also provide a "reference model" on which all other chains will be superposed. If a reference model is not explicitly provided, the first chain found in the list of input files will be used instead. However, it may be advantageous to explicitly define this model if you are concerned about the quality or consistency of the structural alignments.
Homologous structures and insertion codes are supported. To ensure that the residue numbering is interpreted correctly, you will need to uncheck the box labeled "Assume identical chain numbering". The sequence provided as input will be used to pick out similar chains, which will then be aligned to the reference model. The numbering displayed in the output grids will reflect the residue IDs in the reference model; for residues present in other chains but not the reference model, the residue ID will be displayed as "--", but the residue name(s) will still be shown. The primary limitation at this point is the lack of any direct link back to the original residue IDs, but the models displayed in Coot and PyMOL preserve these IDs.
All output files will be written to a directory named StructureComparison_X (where X is the job ID). If structure superpositioning is enabled, there will be a separate PDB file for each chain, plus CCP4 map files if map coefficients were available. If the map coefficients were calculated from input X-ray data (not pre-calculated) these will also be written out.
When the application is finished running, a summary tab will show a list of all chains used for the analysis. The suppary tab also lists basic statistics (such as number of atoms and residues, mean isotropic B-factor, ...). Additional tabs are added for each analysis. Each tab contains a grid with chains organized one per row, and residues of interest in columns. Color coding indicates the relationship of the specific residue in each cell to the equivalent residues, and/or properties such as validation outlier or multiple conformations. Cells colored in grey indicated residues missing from the input chain.
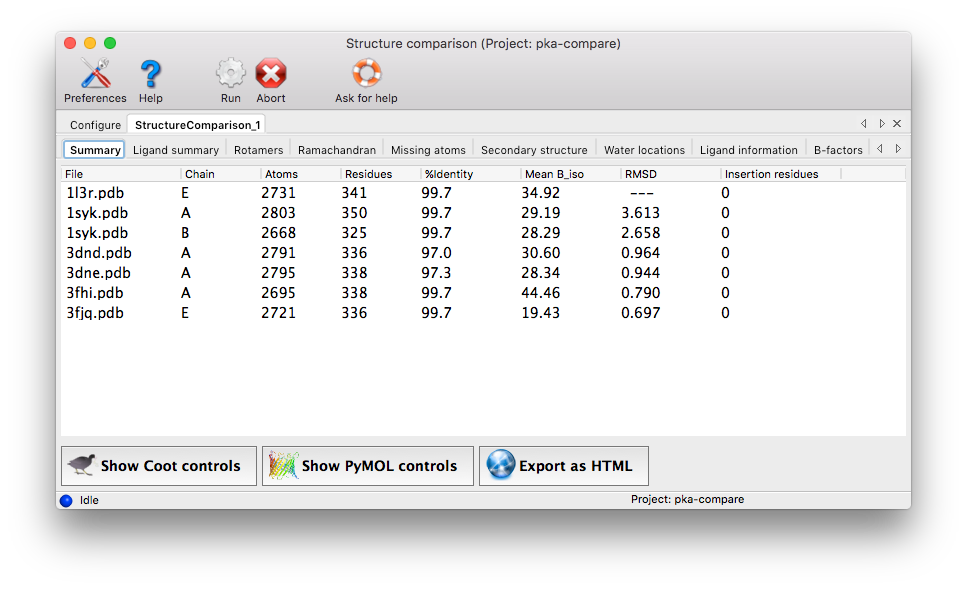
The criteria used to display results vary among result tabs, but as the primary purpose of the program is to highlight regions of difference, homogeneous regions will usually be omitted from the grid.
The following analyses are carried out. It should be noted that the result tabs only appear if differences were found.
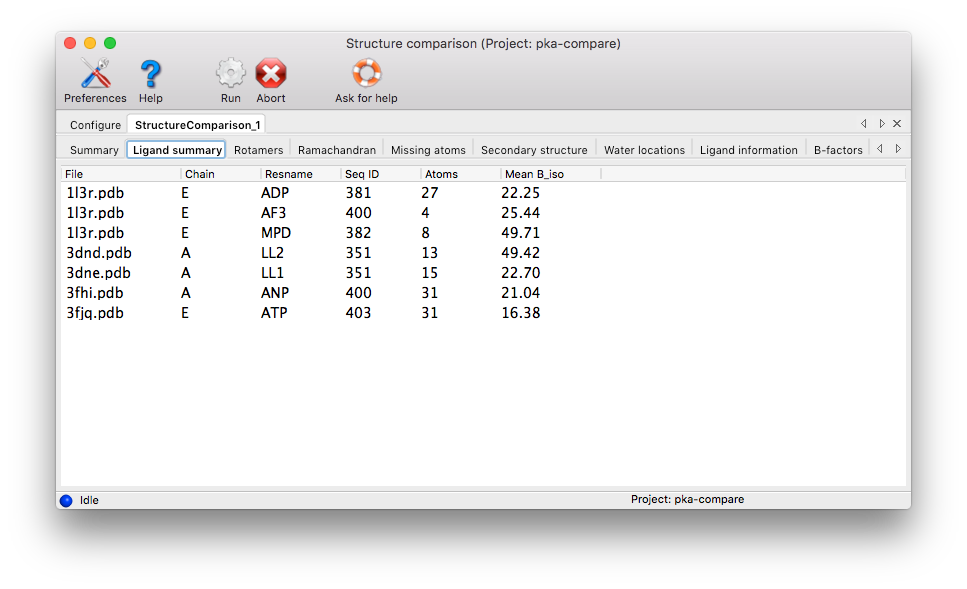
Ligand Summary: If ligands are present in at least one of the chains, they are listed in the results table, along with basic properties such as residue name, number of atoms and mean isotropic B-factor. Note that columns are sortable, f.ex. the table can be sorted according to the number of atoms.
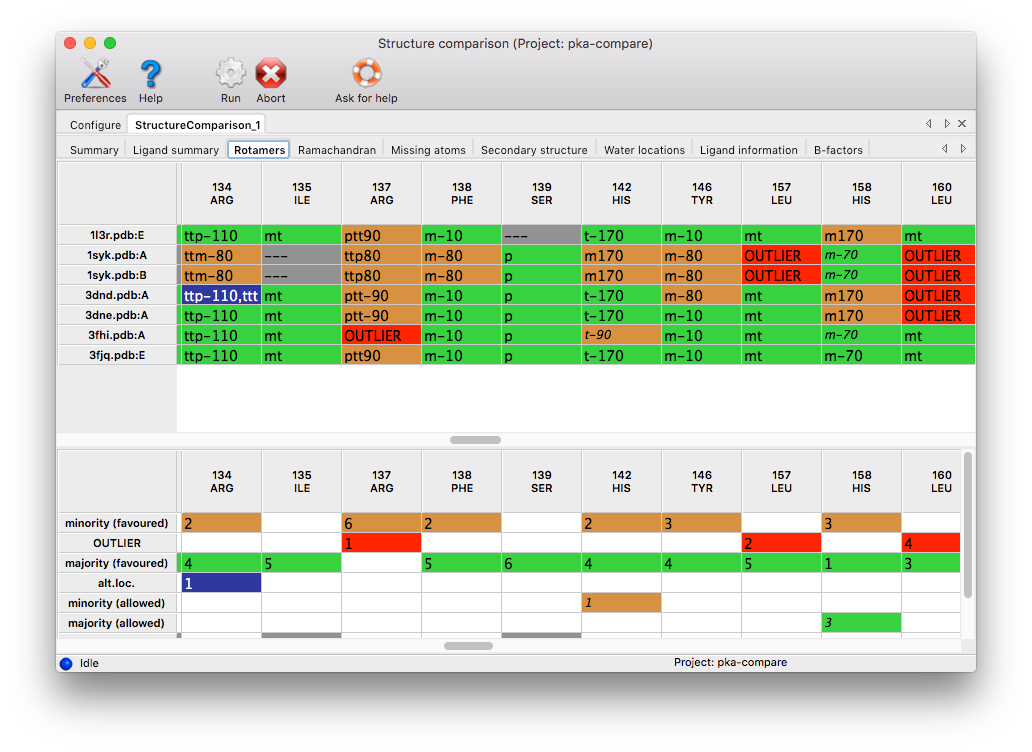
Rotamers: Any set of matching residues containing more than one rotamer type will be included, plus all outliers. Coloring: red for outliers, orange for "minority" conformations, green for "majority" conformations (at least 50% of chains under consideration), blue for multiple conformations, grey for missing side-chains or residues. The rotamer IDs are taken from the Ultimate Rotamer Library, which is described in Hintze et al. (2016). In the bottom part of the window is a summary table which can serve as a legend for the cell colors.
Ramachandran angles: Any set of matching residues with different phi,psi angles plus any outliers will be displayed. Additionally, residues which fall into more than one distinct "favored" region will be included (this is somewhat redundant with the secondary structure comparison). Coloring: red for outliers, orange for "allowed" angles, green for "favored", blue for multiple conformations, grey for missing residues.
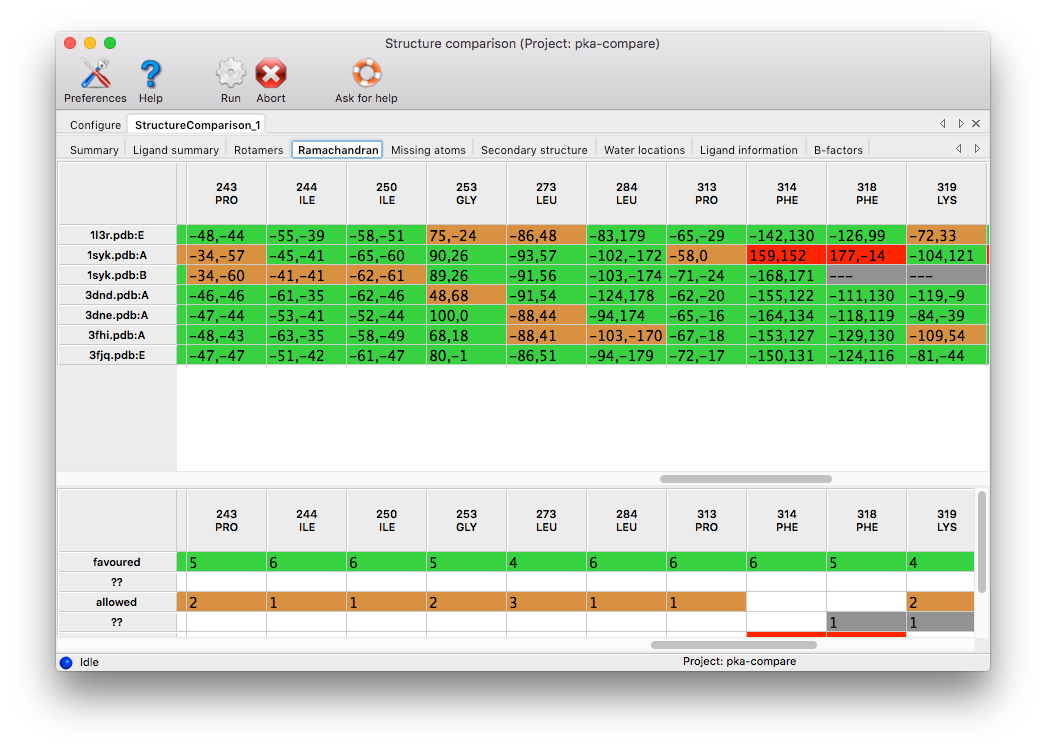
Missing atoms: Any set of matching residues with at least one atom missing will be displayed. Coloring: red for residues missing backbone atoms (C, O, N, CA, or CB), orange for residues missing sidechain atoms, blue for entire missing residues, green for complete residues. The cell width can be expanded to read the list of atom names in the cell.
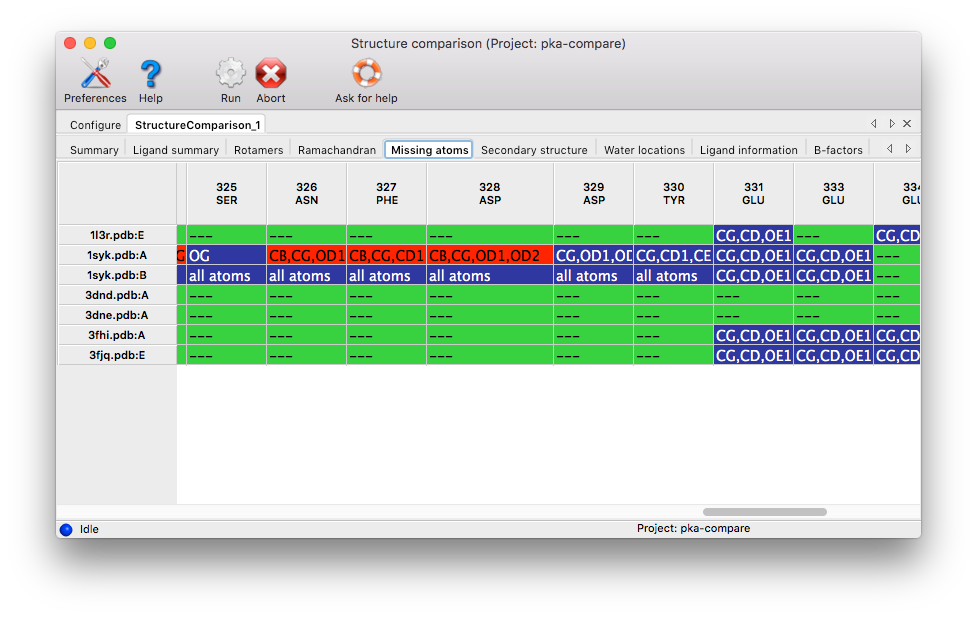
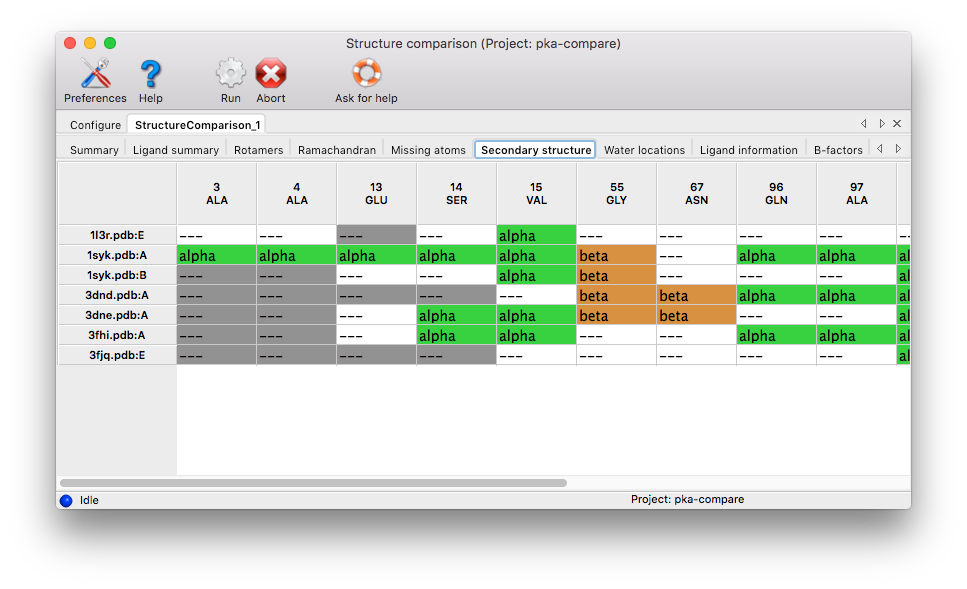
Secondary structure: Any set of matching residues with more than one type of secondary structure assignment will be included. Coloring: white for unstructured/loop, green for alpha helices, orange for beta-sheets, blue for 3_10 helices, grey for missing resdiues.
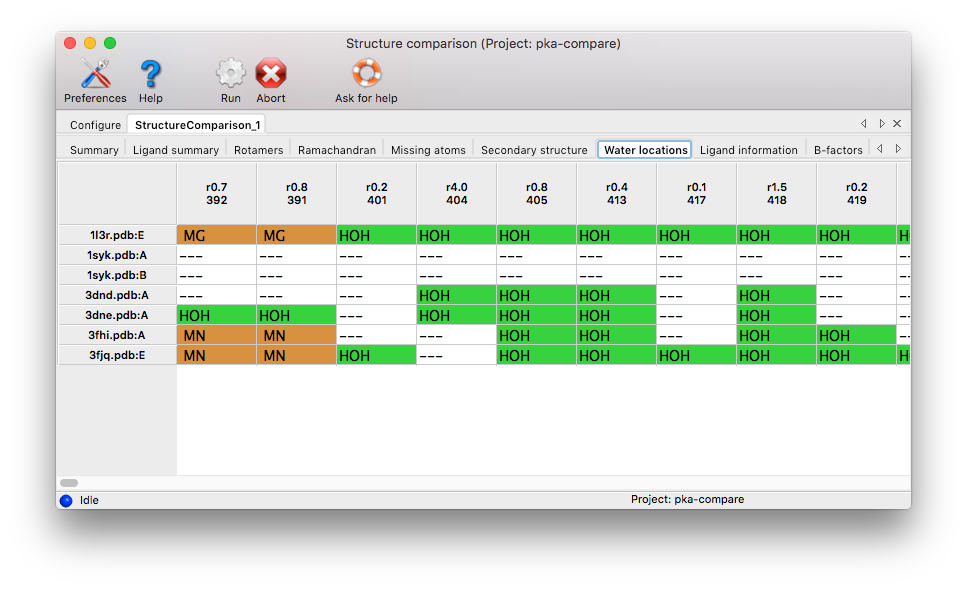
Water locations: Any water molecules not conserved in all chains are listed in the results table. Likewise, ions are included in the analysis. It is of particular interest to investigate areas with waters in some chains and ions at equivalent positions in other chains. Mixed ion/water sites are therefore listed at the leftmost part of the results table. Coloring: orange for ions, green for water molecules, white for sites without corresponding water molecule.
Ligand Information: If ligands are present, their centers of mass are compared. The table is useful to assess if ligands are at equivalent positions in all chains. Coloring: blue for ligands, white for sites without ligand at the equivalent position.
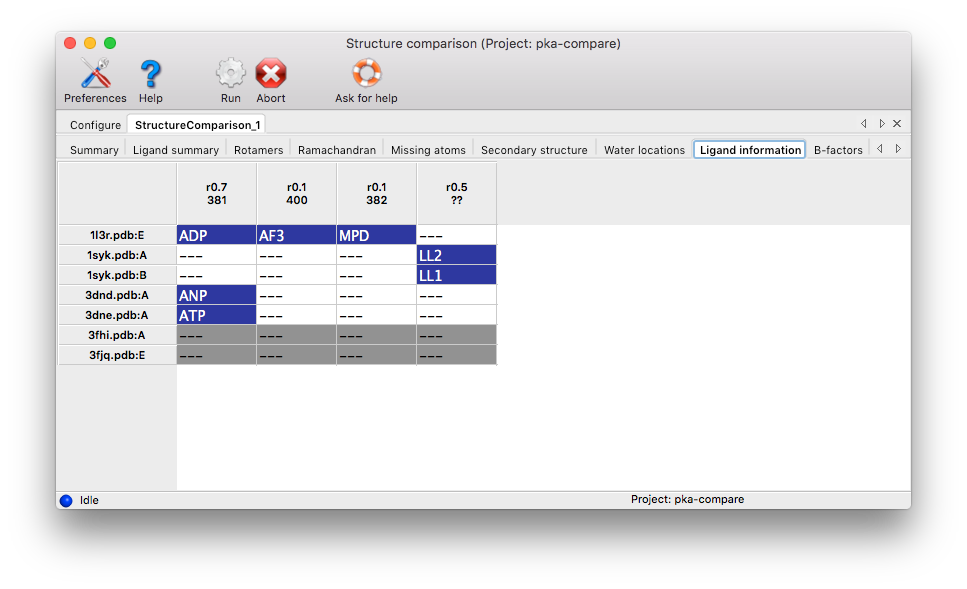
Histidine Protonation: Histidine side chains can have three different protonation states. If H atoms are present in at least one input chain, the results tab lists all histidine residues with different protonation states.
Omega Angles:: Omega angles are categorized as trans, cis or twisted. Any matching residues with different assignments are displayed in the results table.
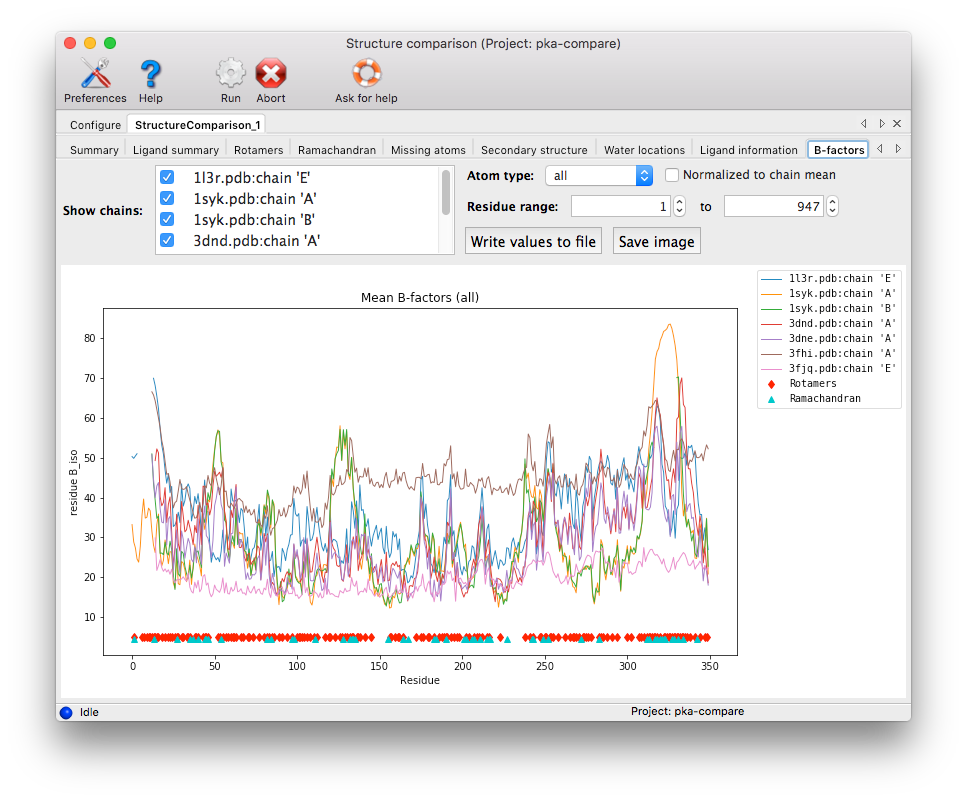
B-factors: Unlike the other analyses, this result does not identify heterogeneity on a per-residue basis, but instead plots the mean B-factor for each residue by chain, with the option of viewing only the main-chain or sidechain average. In addition to the absolute mean values, it can also display the values normalized to the overall mean for each chain, which may be useful when the structures being compared differ significantly in the scale of their B-factors. Points denoting rotamer and Ramachandran outliers will also be displayed. (Note that this plot does not interact directly with Coot or PyMOL.)
To show controls for sending data to and from Coot, click the icon on the toolbar of the results window. A small panel will appear:
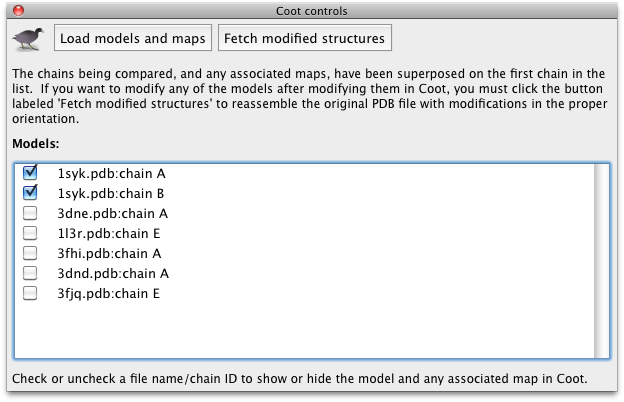
Click the button labeled "Load models and maps" to read all structures into Coot simultaneously. Once this is complete they will appear in a checklist in Phenix; unchecking any structure will hide the associated objects in Coot. As with other validation programs in the Phenix GUI, clicking on the field for any residue in the results grid will recenter Coot on that residue (in the appropriate frame of reference depending on whether models were superposed). You may now adjust the models to match a consensus if desired.
If the structures were superposed, you should not try to use the models in Coot for further refinement. Instead, click the button labeled "Fetch modified structures" to write out the models from Coot to the results directory. Phenix will automatically recover the original orientation and replace the unmodified chain in the input structure with the model from Coot, while leaving all other atoms in place. The complete files will be written out to the result directory, renamed with the extension "_modified.pdb".
Coming soon.
- Hintze BJ, Lewis SM, Richardson JS, Richardson DC. Molprobity’s ultimate rotamer-library distributions for model validation. Proteins-Struct. Funct. Bioinforma. 2016 84:1177–1189. PubMed PMID: 27018641
- Madhusudan, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol. 2002 9:273-7. PubMed PMID: 11896404; PDB ID 1l3r
- Wu J, Yang J, Kannan N, Madhusudan, Xuong NH, Ten Eyck LF, Taylor SS. Crystal structure of the E230Q mutant of cAMP-dependent protein kinase reveals an unexpected apoenzyme conformation and an extended N-terminal A helix. Protein Sci. 2005 14:2871-9. PubMed PMID: 16253959; PDB ID 1syk
- Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science 2005 307:690-6. PubMed PMID: 15692043; PDB ID 3fhi
- Orts J, Tuma J, Reese M, Grimm SK, Monecke P, Bartoschek S, Schiffer A, Wendt KU, Griesinger C, Carlomagno T. Crystallography-independent determination of ligand binding modes. Angew Chem Int Ed Engl. 2008 47:7736-40. PubMed PMID: 18767090; PDB IDs 3dnd, 3dne
- Thompson EE, Kornev AP, Kannan N, Kim C, Ten Eyck LF, Taylor SS. Comparative surface geometry of the protein kinase family. Protein Sci. 2009 18:2016-26. PubMed PMID: 19610074; PDB ID 3fjq