Contents
phenix.superpose_pdbs can superimpose only between pdb files that have equally/similarly aligned nucleic names. This applies to all pdb input files (not only cryo_fitted files). Consider to align both input files by "python <User Phenix Path>/modules/cryo_fit/steps/9_after_cryo_fit/align_nucleic_acid_name_into_middle/align_nucleic_acid_name_into_middle.py"
- If a user input pdb file has unreasonable structural geometry. Although the fit between atomic model and map looks good, it is a fictitious fitting without considering ideal geometry. Run cryo_fit to find decent fit to the cryo-EM map along maintaining reasonable structural geometry.
- Providing higher (better) resolution map tends to help this problem.
- Enforcing stronger map weight tends to help this problem.
- If cryo_fit is provided a giant cryoem map with a tiny atomic model.
- Then, the cryo_fit calculates the gradient of CC because of the large empty space not filled. The constraint forces for the model are not helping as they are very small.
- 3-1. Re-run cryo_fit with an atomic model that fits the majority of the map.
- Fit multiple atomic models into a symmetric map or sequential fitting into a non-symmetric map. Watch https://www.youtube.com/watch?v=6VGYo1pRRZ8&t=0s&list=PLVetO1F9gm_oa--j37yrjzJ4yVJYzPVN5&index=12
3-2. Re-run cryo_fit with only relevant map region. A user can extract relevant map region by phenix.map_box (preferred) or phenix.map_to_model
- If the initial model is not properly aligned to a map, fit using UCSF Chimera -> Tools -> Volume Data -> Fit in Map
- Open pdb file
- [menu]
- Select -> Residue -> HIS
- Tools -> Structure Editing -> Rotamers -> OK
- (select the most probable rotamer each)
- File -> Save PDB
"I edited out lipids, HEM and other hetero atoms and I verified that they are all gone. However, still my pdb file is not clean enough for gromacs_cryo_fit".
Based on Doo Nam's experiences, this error message may indicate between two cases/scenarios.
(The 1st case) It means that atomic model is not stable enough. When Doo Nam ran phenix.real_space_refine and provided real_space_refined pdb file into cryo_fit, the problem was solved.
(The 2nd case) It means that the map dimensions need to be larger. Therefore, check map size and solve a problem with VMD/EMAN2/relion like followings.
Like other MD simulations, gromacs need enough map box size to cover the atomic model to run (ziggle and wiggle). Refer Waters seems to be out of the box
For example, stuck-out red oxygen atoms outside the right edge of the box are the problem.
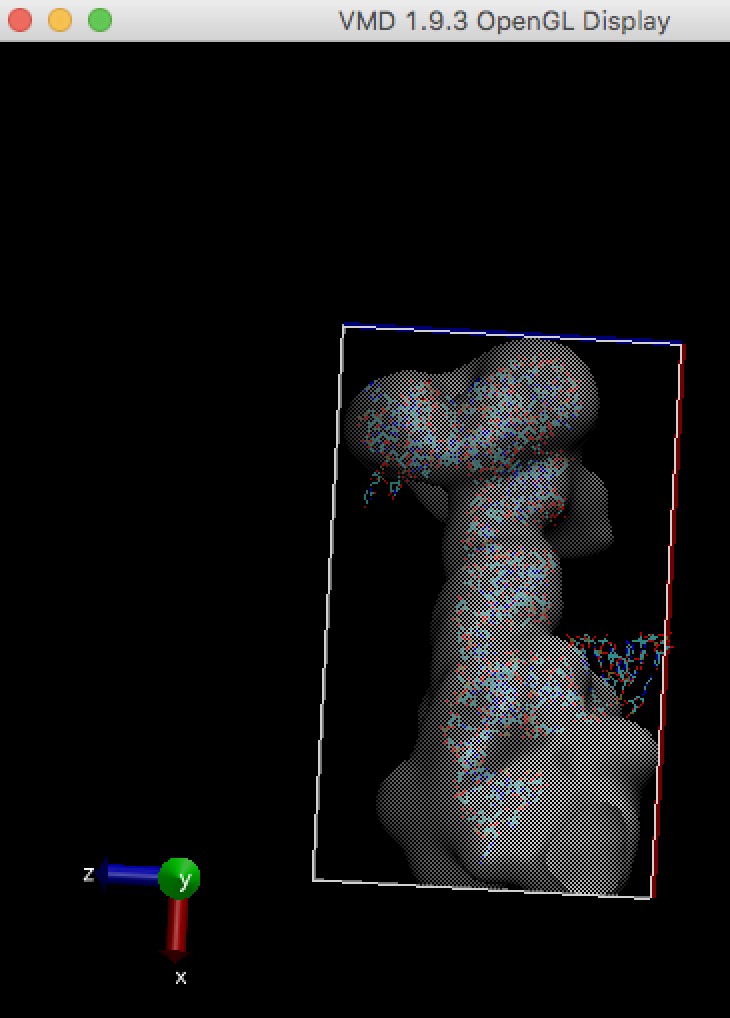
In order to run any MD simulation (including cryo_fit), a box should be large enough like
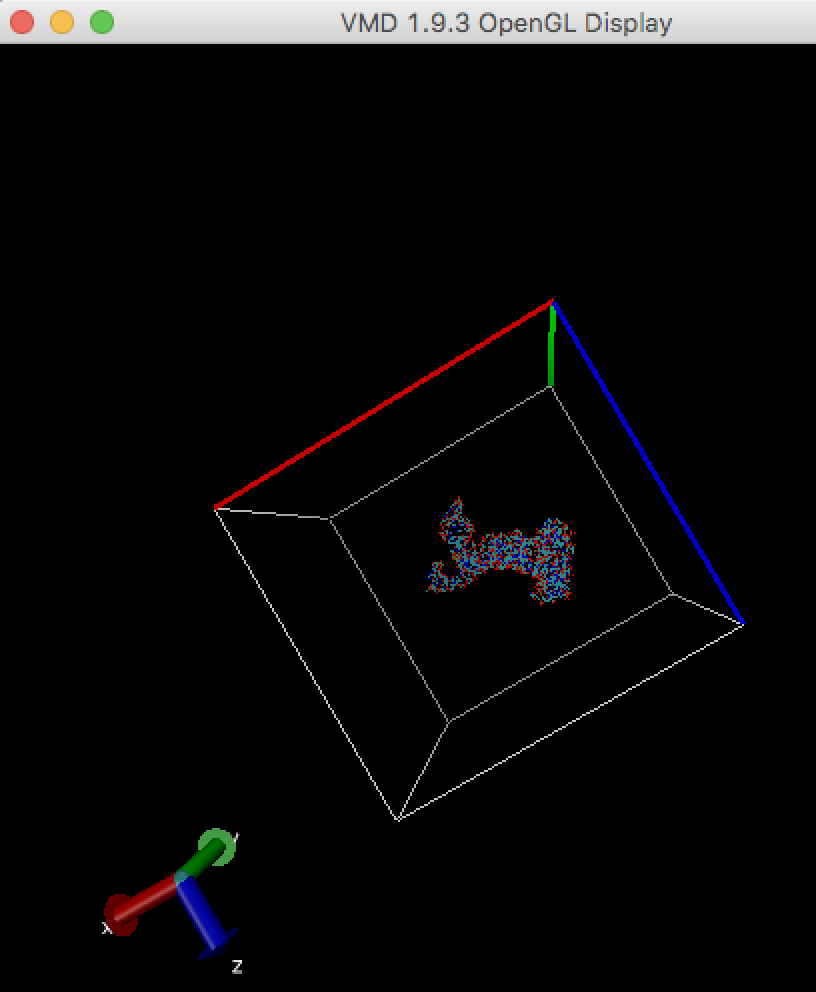
Make map box size larger (see "How to enlarge map box size?" in this FAQ), and run cryo_fit again. You can check map box size by VMD. Alternatively, remove sticking out atoms if these are unnecessary, then run cryo_fit again.
For protein modeling, I would use cryo_fit2 which is not limited by box size requirement. Most of the time, it better fits than cryo_fit1 in terms of fitting and geometry preservation anyway.