| Python-based Hierarchical ENvironment for Integrated Xtallography |
| Documentation Home |
The phenix.refine graphical interface
GUI overviewThe graphical interface for phenix.refine runs the unmodified command-line version; default settings are unchanged. All parameters should be configurable through the GUI, although some of these (such as NCS restraint groups) are handled in a non-standard way. This document describes the behavior of the GUI only; see the phenix.refine documentation for details on specific parameters and methods. At present the GUI has the following capabilities:
ConfigurationBecause phenix.refine currently has nearly 500 distinct parameters, many of which rarely need changing, and the window controls are mostly created automatically, advanced settings are hidden by default. The "user level" may be set within each window, or set globally in the Preferences. The most important parameters (input and output files, strategy, additional procedures such as solvent updating and simulated annealing) are shown in the main window. Input filesThe first tab contains settings for input files, which can be added by clicking the "+" button below the file list, or by dragging files into the list control and releasing the mouse button. Up to five different reflection files may be used (X-ray data, X-ray R-free flags, neutron data, neutron R-free flags, and experimental phases), but in most cases all data will be in a single file. Any number of PDB files for the starting model may be added; these will be combined internally. You may also specify a reference model for additional restraints (see below). Other supported file formats are CIF (restraint) files for non-standard ligands, and phenix.refine parameter files. 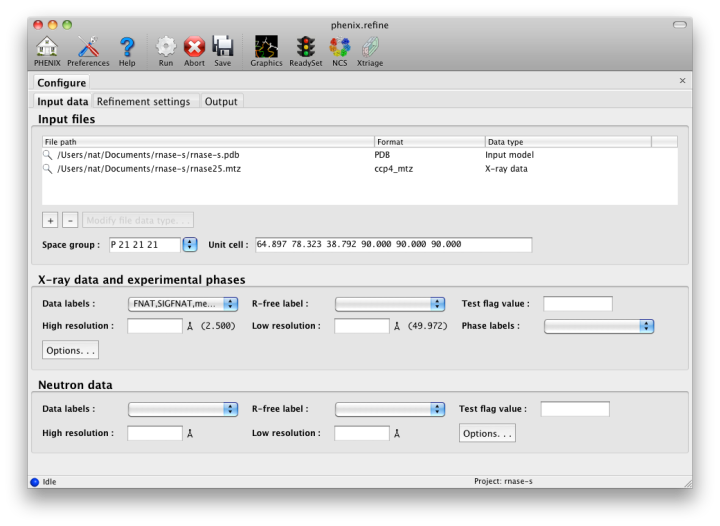 The GUI will attempt to guess the purpose of PDB and X-ray files, but you may change this by right-clicking on the "Data type" field associated with the file and selecting an option from a drop-down menu, or select the file and click the button labeled "Modify file data type". If your reflection and/or PDB files have the appropriate information on crystal symmetry and data labels, these parameters will be extracted automatically. Appropriate column labels for reflection files will also be added to the drop-down menus; by default, anomalous intensities (I+/I-) will be used if present, but you may also use merged data and/or amplitudes. The R-free flags are optional, since phenix.refine will add them automatically if none are present, but once flags have been used in refinement (including the automated building in AutoSol or AutoBuild) the same set should always be used. Refinement settingsThe second tab contains options for the refinement protocol. Many of these controls will open additional dialog windows in response to mouse clicks or file selection. In many cases, you can open these windows on demand by right-clicking on the appropriate control (for instance, right-clicking the "Rigid body" strategy button will show the atom selection controls for defining rigid groups). All other parameters can be found in the Settings menu. 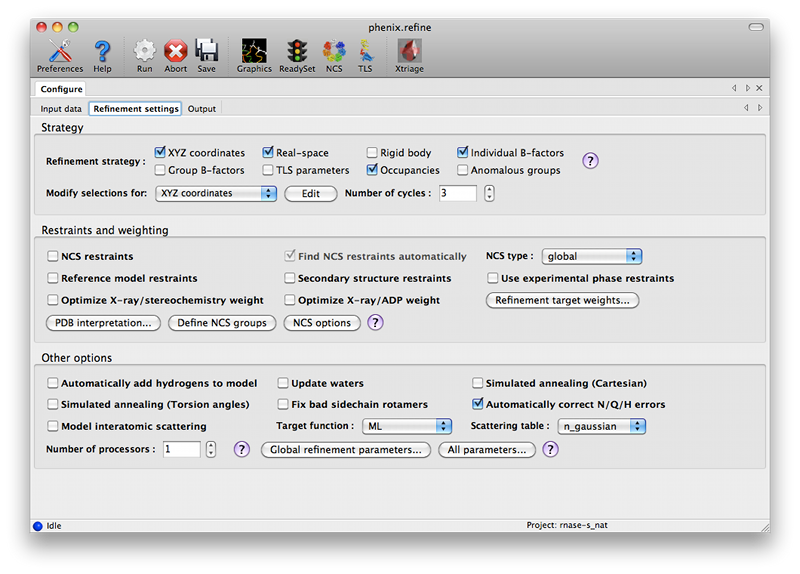 Most of the refinement options presented on this tab essentially fall into three categories:
The default options have been chosen to work well for partially refined structures at moderate resolutions, which comprise the majority of structures in the Protein Data Bank. However, at significantly higher or lower resolution, or for structures at the early or final stages of refinement, additional options may be necessary or beneficial. The first set of controls determines the parameterization of the model and the attributes to be refined. By default, the Individual Sites, Individual ADPs (B-factors), and Occupancies strategies are selected, since these are usually appropriate at a wide range of resolutions and refinement stages. All strategies have associated atom selections which control what parts of the model are refined. A brief summary follows; note that the resolution limits specified are approximate only, and in practice may vary widely depending on model quality, data quality, and observation-to-parameters ratio.
The refinement protocol can be further modified by manipulating various restraints (structural and otherwise):
Several other options are available for the types of optimization used; these typically apply globally, except as noted.
The output tab allows you to change the destination directory and modify the default map coefficients and/or map files written out. By default, an MTZ file containing 2mFo-DFc and mFo-DFc map coefficients will be generated. 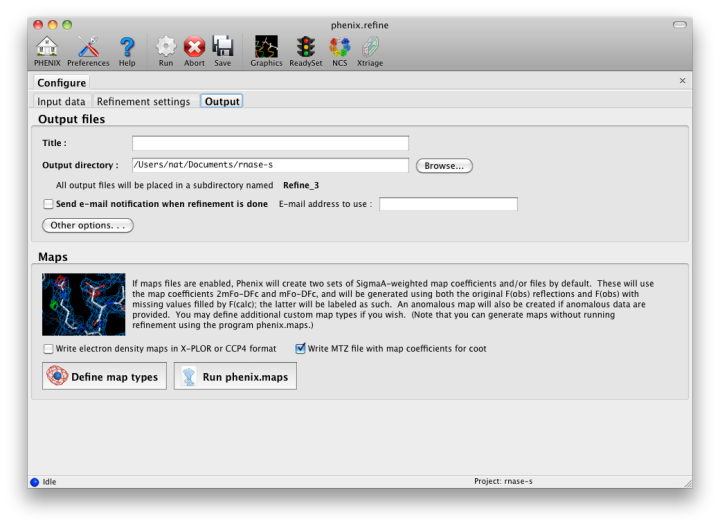 Creating restraintsIf the PDB file contains ligands or prosthetic groups that are not part of the limited monomer library distributed with PHENIX, a warning message will be displayed when the file is loaded. The necessary restraints must be generated by phenix.elbow in order for refinement to proceed. To start this process, click the ReadySet button in the toolbar or choose "Prepare structure and restraints" from the Utilities menu. This will launch a dialog for running phenix.ready_set, a simple utility for preparing structures for phenix.refine. 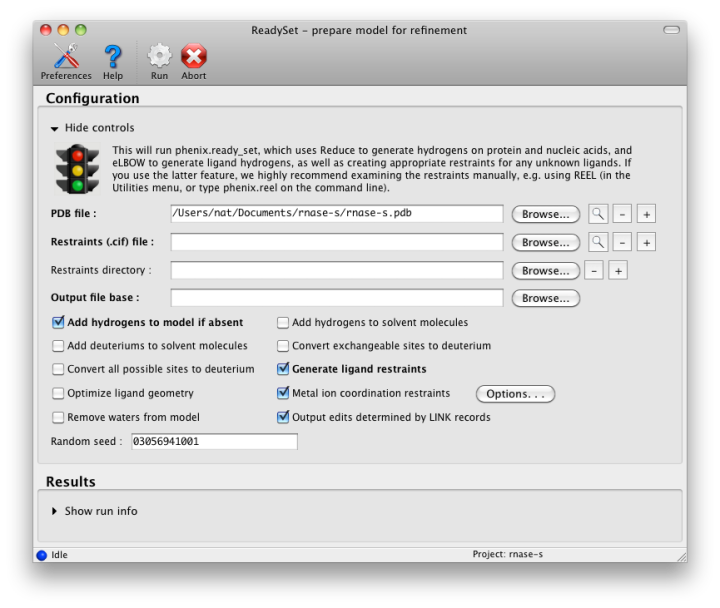 Generating the restraints will take between 30 seconds and several minutes, depending on computer speed and the size of the ligand. When ReadySet is finished, another window will be displayed summarizing the results. If a project directory is defined, these can be saved for future runs and will be automatically loaded into the GUI when launched. If your input model does not contain hydrogen atoms and you would like to use them in refinement, this can also be done via the ReadySet dialog, or by clicking the box labeled "Automatically add hydrogens to model" in the main window. Note: ReadySet look in the PDB's Chemical Components Database (distributed with PHENIX) for the three-letter code of any unknown ligands it finds, and use the database information to generate restraints. For non-standard ligands, users are advised to run eLBOW with a SMILES string or equivalent source of information. Generating robust geometry restraints from a PDB file alone is problematic, because the coordinates are not a reliable guide to molecular topology, chirality, etc. Neutron crystallographyphenix.refine can perform refinement using either X-ray data, neutron diffraction data, or both. You can change the type of data contained in a reflections file by right-clicking on the "Data type" field in the file list. At present, phenix.refine always requires that you define an X-ray dataset; this constraint will be removed in future versions. If you are performing refinement against neutron data alone, you must treat the reflections as if they were X-ray data. All of the data options remain the same, and will be extracted automatically. You must change the scattering table to "neutron" in the "Refinement settings" tabe. If you are doing joint X-ray/neutron refinement, you should leave the scattering table set to the default, and specify one reflections file as neutron data. Parameters for neutron data are not chosen automatically; once you have specified a neutron reflections file, click the "Neutron data. . ." button to select the column labels for the data and test set. If you need to add hydrogens and deuteriums to your model before running phenix.refine, you can either run ReadySet prior to refinement, or change the options for automatic hydrogen addition. If your model already includes these atoms, you may wish to turn automatic hydrogen addition off to ensure that existing atom types are left in place. Atom selectionThe atom-selection syntax used by PHENIX is described in the phenix.refine manual. In the GUI, any valid atom selection can be visualized if you have a suitable graphics card and have already loaded a PDB file with valid symmetry information. The graphics window can be opened by clicking the "View/pick" button next to any atom selection field. Depending on the size of your structure, it may take several seconds for PHENIX to determine the atomic connectivity. The current selection, if any, will be highlighted: 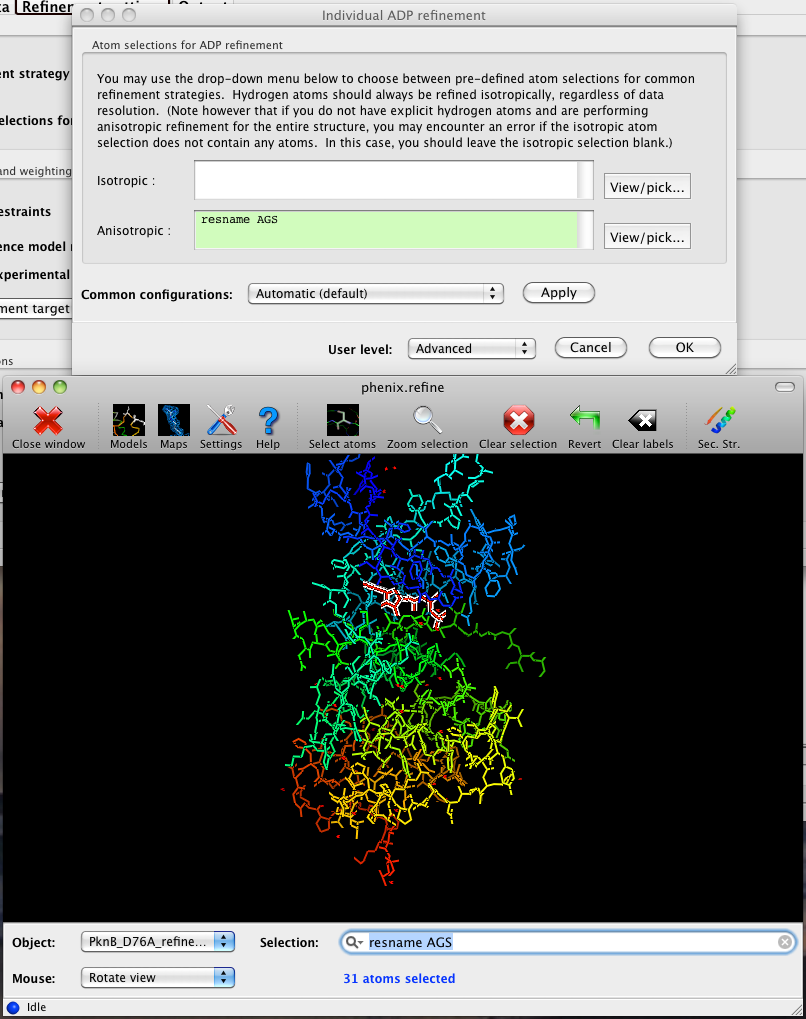 The Select atoms button opens a window that allows you to type in selections and immediately visualize the results, including the number of atoms selected. On mice with at least two buttons, clicking on an atom with the right button will open a menu for selecting residue ranges or chains. However, we recommend that you learn the selection syntax, as it is much more flexible than mouse controls. Selections made in the graphics window will be sent automatically to the appropriate control. Once this window is open, it does not need to be closed; clicking a different "View/pick" button will transfer control of the display. (On Linux, you will first need to open the Actions menu and click Restore parent window to transfer mouse and keyboard controls back to the configuration dialog.) Identifying and editing TLS groupsPHENIX includes a tool (phenix.find_tls_groups) for fast identification of appropriate TLS groupings based on isotropic ADPs in a PDB file. This is integrated into the phenix.refine GUI, and can be accessed from the TLS group editor (the "TLS" button on the toolbar, also available on the "Refinement settings" configuration tab). The editor is essentially just an atom selection list control similar to others in PHENIX: 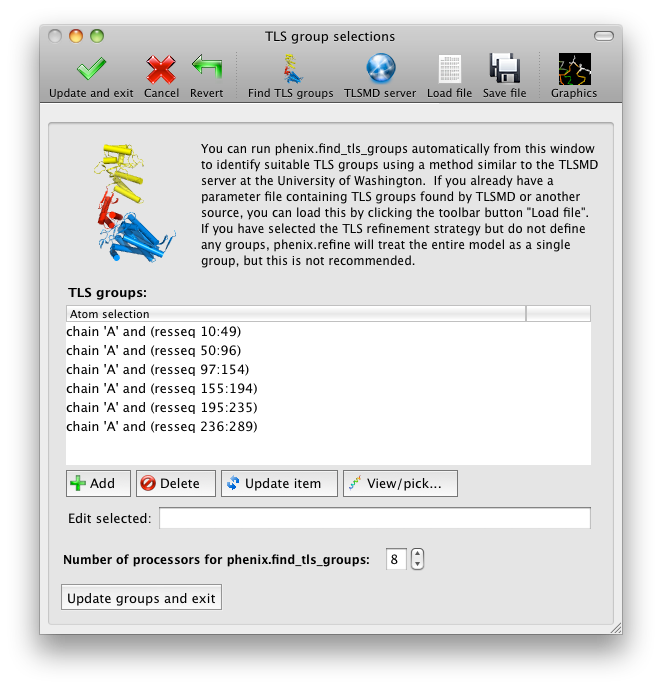 Clicking the "Find TLS groups" button will launch the identification program, which typically takes a few seconds to a few minutes to run depending on the size of the structure and the number of processors available. All CPU cores will be used by default, but this can be adjusted using the control near the bottom of the window. You may alternately specify TLS groups manually, or upload a file from the TLSMD server. Once you have the desired groups input, close the editor by clicking either of the buttons at the upper or lower left of the window. Running phenix.refineEach time phenix.refine is run from the GUI, a new folder is created (in the current output directory) named Refine_X, where X is an integer corresponding to the job ID. This directory will contain all temporary and output files created during the course of the run. Existing folders will not be overwritten. All output files will be displayed in the GUI in the results tab. Once you are done configuring refinement, switch to the Run tab in the main window and start the process by clicking the "Run" button. The log output will appear at the bottom of the new tab: 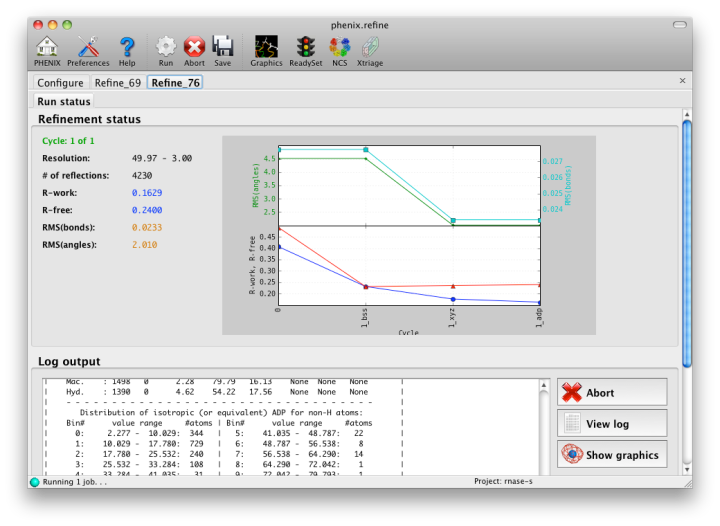 Statistics will be continually updated; any that appear to be outliers (e.g. excessively high R-factors) will be highlighted in red. If an error occurs during refinement, a message box will pop up and the process will be halted. After refinementOnce the refinement is complete, additional tabs will appear. The first of these is a summary page displaying the final statistics, a list of output files, buttons for opening the refined structure in Coot and PyMOL, and links to assorted graphs. Note that the output files may include one ending in "_data.mtz"; if you did not specify R-free flags as part of the input, the new file will contain the automatically generated flags, and should be re-used in future runs. 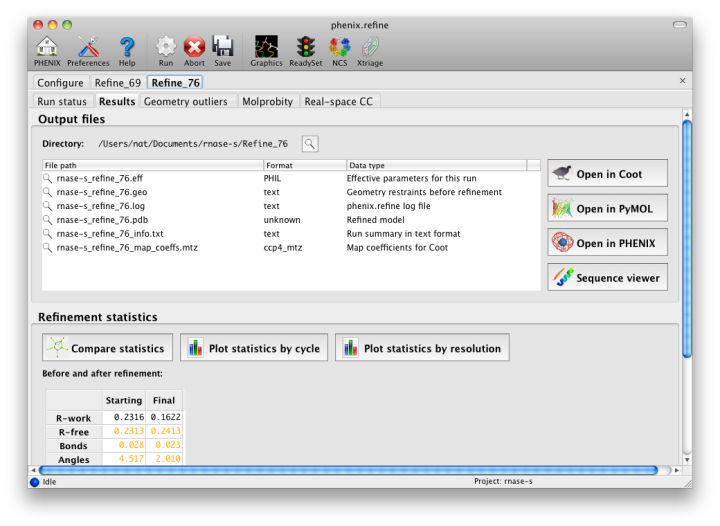 Additional tabs contain the full validation summary, which is essentially identical to the standalone validation program. Frequently Asked QuestionsQuestions not specific to the graphical interface are answered here.
Acknowledgementsphenix.refine was primarily written by Pavel Afonine and Ralf Grosse-Kunstleve, with additional contributions from Peter Zwart, Jeff Headd, Nat Echols, Nigel Moriarty, and Tom Terwilliger. The electron-density isosurface and anisotropic ellipsoid code in the CCTBX (used in the internal graphics viewer) was contributed by Luc Bourhis. References
| |